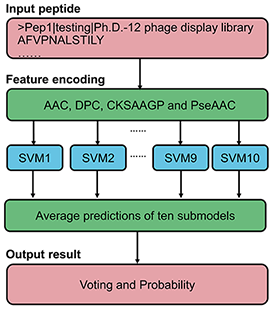
PDL1Binder is an ensemble support vector machine (SVM) based predictor for identifying PD-L1 binding peptides. The framework of PDL1Binder is illustrated in the following figure. For a given peptide, it will be predicted by ten submodels separately. PDL1Binder then uses the averaging voting method and makes final prediction based on the average probability value.
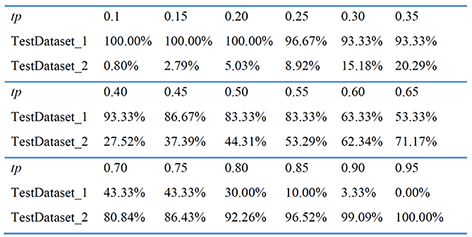
PDL1Binder allows users to set the threshold of probability value to differentiate between predicted positives and negatives (tp). Take the two independent testing datasets of "Download" page as an example, the model performance at different thresholds is shown in the Table below.
PDL1Binder allows users to submit peptide sequences in fasta or plain text format. Predictive results of PDL1Binder are displayed in a table.
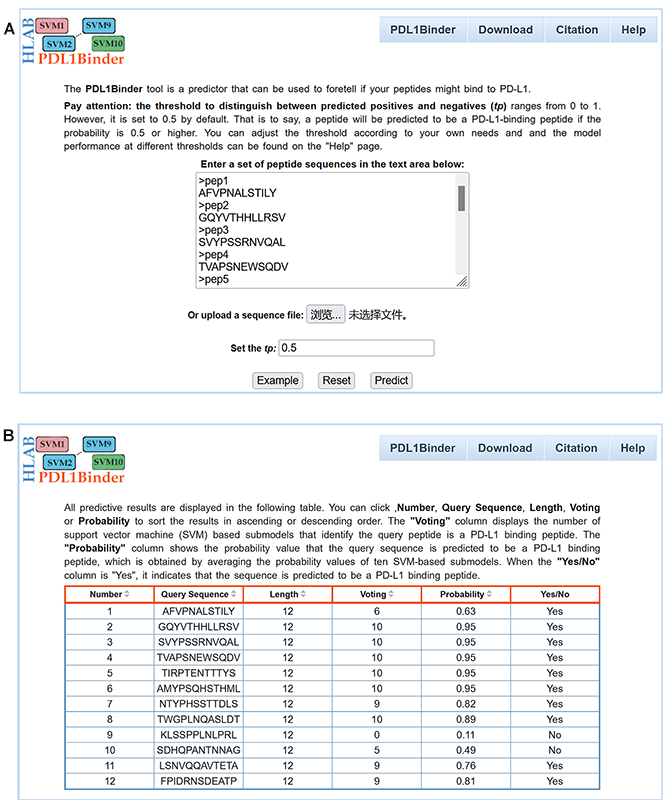
Number: the serial number of the query sequence;
Query Sequence: the sequence of the query peptide;
Length: the length of the query sequence;
Voting: the number of SVM-based submodels that identify the query peptide is a PD-L1 binding peptide;
Probability: the probability value that the query sequence is predicted to be a PD-L1 binding peptide, which is obtained by averaging the probability values of ten SVM-based submodels;
Yes/No: the "Yes/No" column shows the prediction result, when the "Probability" is greater than or equal to "tp", the column is displayed as "Yes", which indicates that the sequence is predicted to be a PD-L1 binding peptide; otherwise "No".