

MIMOX is a web tool for bioinformatical analysis of peptide sequences derived from phage display. It has two sections. The first section utilizes ClustalW to align a set of mimotopes and then deduce consensus sequence based on the aligned mimotopes through a simple statistical method or through JalView. The second section searches for possible reasonable explanations for the consensus sequence deduced. It maps the consensus sequence back to its corresponding PDB structure, finds out possible spatial neighbor clusters that the consensus sequence might stand for, utilizes NACCESS to evaluate candidate neighbor clusters and Jmol to view them in 3D context. The name MIMOX is related to the term "mimotope". You can take MIMOX as an abbreviation for MIMOtope eXplorer or MIMOtope eXplanation? Whatever, we just call this web application MIMOX.
As early as 30 years ago, Atassi and his colleagues (Biochem J 1976, 159(1): 89-93. JBC 1977, 252(24): 8784-8787.) showed that an antibody-combining site could be mimicked synthetically. In another word, they suggested that conformational epitope can be simulated by synthetic peptide. 20 years ago, the term "mimotope" was introduced by Geysen and his colleagues (Mol Immunol 1986, 23: 709-715.). In that paper, Geysen stated "A mimotope is defined as a molecule able to bind to the antigen combining site of an antibody molecule, not necessarily identical with the epitope inducing the antibody, but an acceptable mimic of the essential features of the epitope".
All mimotopes are antigenic mimics of native epitope, because they bind to the same antibody competitively. Some mimotopes are even immunogenic mimics of native epitope. That is antibody produced by "immunogenic mimics" immunization will also cross react with natural antigen. Mimotopes are useful. For example, antigenic mimics can be used as diagnostics if they are selected out by disease-specific antibody. And immunogenic mimics might be good candidates for new vaccine development. Besides, the sequences information of a set of mimotopes from a given experiment might be useful for mapping the corresponding natural epitope or at least providing some useful hints to understand the natural epitope further. However, as mimotopes were identified and obtained from synthetic peptides initially (as both Atassi and Geysen group did), once it was very hard to get one mimotope. The situation has been greatly changed since George P Smith developed the phage display technique (Science 1985, 228(4705): 1315-1317.).

George P Smith:
the father of phage display
Phages are viruses that infect bacteria. Some phages are good expression vectors that can express foreign DNA sequence inserted in their coat protein genes. When the hybrid coat protein is packed into phage particles, the foreign peptide is displayed on the outer surface. A phage display library is a heterogeneous mixture of such phage clones, each carrying a different foreign DNA insert and therefore displaying a different peptide on its surface. The most common type is random peptides phage display library. Because the foreign peptides or random peptides are displayed on the surface, it is easy to select or screen for special phages with antibody, receptor, enzyme, and other selectors. The selected phage is easy to replicate in bacteria. In fact, phage display technique constructs a rapid in vitro evolving system with mutation, replication, and selection mechanism. A schematic diagram of phage display is shown below. For more information, we recommend you read the review Phage Display written by George P Smith and his colleague though it was published almost 10 years ago.
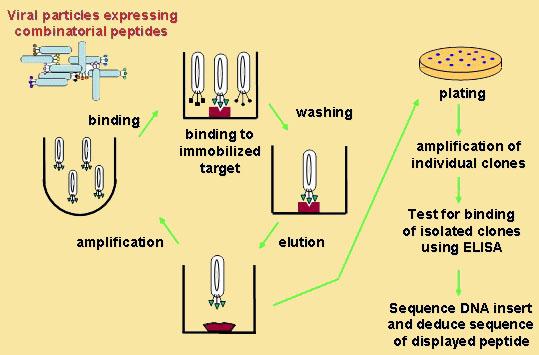
Scheme of phage display
The impulse to develop this web service originated several months ago when we worked on a conformational epitope database. Many research reports have shown that phage display is also a promising method for epitope mapping. Years ago, some groups had to analyze their mimotopes manually. Recently, a few kinds of software for phage display analysis have been developed. Most of the existing tools have not been implemented as an online service until now however, making it less convenient for the community to access, utilize, and evaluate them. Why don't we make one? It should be in English, no necessary to set up, independent of operating system, freely available and accessible via internet. So the web tool with the name MIMOX came into our mind.
MIMOX is a free online service powered by Apache. It is a CGI program coded with Perl. Some Bioperl modules are used when coding. This web application also utilize other exist programs such as CLUSTALW, JalView, NACCESS, and Jmol to implement corresponding function directly. The algorithm of MIMOX is simple. It is assumed that residues of common sequence deduced from phage display peptides and residues of the native epitope have similar physicochemical properties and spatial relationships since they bind to the same antibody competitively. As the residues and their arrangement of common sequence is known or deduced from multiple alignments of mimotopes, MIMOX search all candidates that have same or similar residues and spatial relationships within the PDB structure of corresponding natural antigen.
As far as we know, there are at least 5 similar programs (or algorithms) that have been used in phage display based epitope mapping. They are SiteLight, FINDMAP, 3DEX, MIMOP, and an unnamed algorithm described by Enshell-Seijffers et al.
The algorithm of SiteLight (by Halperin et al, 2003) has 3 main stages: (1) divides native protein surface into overlapping patches based on geodesic distances between residues; (2) aligns each mimotope in library with each patch and scores and sorts them; (3)iteratively selects high scoring matches until 25% of the native protein is covered. SiteLight was implemented in C++ and has been tested on Red-Hat Linux 7.1. FINDMAP (by Mumey et al, 2003) align a probe (e.g. a consensus sequence of mimotopes) to target (e.g. sequence of native protein), allowing any permutation of the probe sequence. It uses a two-part scoring system to evaluate the quality of alignments and a branch-and-bound algorithm to find an alignment with maximum score. It has been implemented as a C++ program. Enshell-Seijffers et al also described their unnamed algorithm for epitope prediction based on mimotopes on Journal of Molecular Biology in 2003. At first, they convert a set of mimotope sequences into a set of overlapping residue pairs. Then calculate and rank the pair occurrences to get a set of major statistically significant pairs (SSP). Lastly, search them within 3D structure of native protein and link SSP into clusters on protein surface. It is unspecified if this algorithm was implemented. 3DEX (by Schreiber et al, 2005), firstly, divides a mimotope sequence into a set of overlapping subsequences with a user-defined length (3~maximum length of mimotope). Then, it search matching residues of each position of above subsequence against sequence or PDB structure of native protein and link the neighbors iteratively until the first subsequence is complete. Then repeat for the following subsequences to complete the mimotope finally and return the result. It is inferred that 3DEX was implemented in Visual Basic and only run on Windows. However, we met problems to just set up it. MIMOP (by Moreau et al, 2006), which published very recently, includes two approaches. MimAlign predicts potential epitopic regions (PER) from mimotope and native protein sequences and 3D structure (if available). MimCons predicts PER from mimotope sequences but requires native protein 3D structure.
Though above similar tools existed, none of them provide online service. As a free CGI program, MIMOX will sweep off this inconvenience.
You are definitely right. Find some test data here and just try it, you will find it is really quite easy. As a free online service, you can use MIMOX wherever and whenever you connect to internet. You do not need to register or login. You do not need to download any program or client plug-in to set up anything. You do not need to switch your MacOS, Linux, Solaris or any other operating systems to Windows. MIMOX has a concise web interface. All you need to do is to select or input some parameters to run it. For more information, please go to help page.
First, like all other computer programs, bugs might have hidden in it. We hope users send their feedback to help us to catch and fix bugs, to improve and extend the function of MIMOX. Second, we would like to implement all other existed algorithms such as FINDMAP and SiteLight as web applications and hyperlinked them to MIMOX. Thus the users could have more choices and we can do some comparative studies on different algorithms too. To systematically evaluate performance of different algorithms and to develop better new tools, a more complete data set should be constructed at first. Though there are some data in our CED, the phage display entries are limited and the database structure and data content of CED is not quite fit for this purpose. There is also another data source named ASPD. However, it is not a relational database and has not been updated since 2002. So, we would like to develop another database for mimotopes to complement it too.
HLAB | Center of Bioinformatics | UESTC | Chengdu, 610054, China [Feedback]