Help Contents:
The hmmscan search algorithm
The hmmscan is used to search protein sequences against collections of protein profiles, e.g. Pfam. HMMCAS uses hmmscan homology search algorithm in HMMER3.1 which downloads from http://hmmer.org/ to search against the Cas protein family hidden Markov models (HMMs) deposited in the TIGRFAMs and Pfam protein families databases.
Cas protein family hidden Markov models
We first downloaded the multiple sequence alignments or seed alignments for protein families from the TIGRFAMs (ftp://ftp.jcvi.org/pub/data/TIGRFAMs/, version 15.0) and Pfam (ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/, version 30.0) databases, searched the seed alignments with the "CRISPR" keyword and found 139 CRISPR-associated seed alignments in total: 101 from TIGRFAMs and 38 from Pfam. We also downloaded 24 novel Cas families seed alignments from http://omics.informatics.indiana.edu/mg/CAS/. Then, hmmbuild program was used to construct profile HMMs from multiple sequence alignments with default parameters. Table 1 shows the basic information about HMMs of core proteins (Cas1-Cas10, Csf1 and Cpf1) of CRISPR-Cas systems.
Click here to download Table1.
Search Parameters
Default searches
The respective profile HMMs defined thresholds (gathering thresholds) are used to determine hit significance.
Filter: Bias composition filtering on
Input Data
Only one-letter codes for 20 common amino acids and X letter are accepted, and any other characters will lead to an illegal character feedback. The input can be one or multiple proteins, or a complete proteome from bacteria or archaea of your interest. Users are proposed to submit a complete proteome in order to make full use of this website.
Input Examples
We have provided an example file (NC_016025.faa) on the homepage, which has been chosen to show a result set that demonstrates the features available on the results pages.
Search Parameters
Required: The library of Cas protein family HMMs, E-value Cut-off
Optional: Bit score Cut-off, Gathering Threshold, No Bias Filter
Cut-Off Thresholds
The hmmscan algorithm has the ability to set two different categories of cut-offs: significance and reporting thresholds. These cut-offs can be defined either as E-values or bit scores. When setting either category of threshold, there are two values for each of the threshold categories: sequence and hit. A query can match a target in multiple places, defined as a hit (or domain) score. The sum of all hits on the sequence is the sequence score.
For example, trying to match repeating motifs can often be difficult, due to sequence variation in the repeating sequence motif. However, it can be possible to capture all examples of the motif, by relaxing the hit parameter while maintaining a stringent sequence parameter. This means that multiple matches, even if they are not strong matches, can be detected, but the sum of these matches must be sufficient to achieve the sequence score, thereby limiting the rate of false positives.
Significance Thresholds
Significance (or inclusion) thresholds are stricter than reporting thresholds and take precedence over them. These determine whether a sequence/hit is significant or not.
Significance E-values
Sequence and hit significance E-value thresholds will set matches with E-values less than or equal to the cut-off E-value as being significant. The default sequence E-value threshold is 0.01 and 0.03 for hits.
| Description | Sequence E-value threshold | Hit E-value threshold |
|---|---|---|
| Accepted Values | 0 < x ≤ 10 | 0 < x ≤ 10 |
| Default | 0.01 | 0.03 |
Significance bit scores
Alternatively, the sequence and hit significance thresholds can be specified as bit scores. Any sequence or hit scoring greater than or equal to that given threshold will be considered a significant hit. By default, the form on the website is filled with typical values, with the sequence cut-off set to 25.0 bits and the bit cut-off set to 22.0 bits.
| Description | Sequence bit score threshold | Hit bit score threshold |
|---|---|---|
| Accepted Values | x > 0 | x > 0 |
| Default | 25.0 | 22.0 |
Reporting thresholds
The reporting thresholds controls how many matches that fall below the significance threshold are still shown in the results (i.e. reported). As every target model (Cas protein family hidden Markov models) in the library is compared to the query, if all matches were reported, then potentially more outputs would be generated. However, it can often be useful to view borderline matches as they may reveal more distant potential informative similarities to the model. As with the significance thresholds, there is a value for both the sequence and the hit, which again can be defined as either an E-value or a bit score.
Reporting E-values
Any sequence or hit with an E-value less than or equal to that given threshold will be reported. By default, the reporting sequence E-value will be set to 1 and the hit cut-off set to 1.
| Description | Sequence E-value threshold (reporting) | Hit E-value threshold (reporting) |
|---|---|---|
| Accepted Values | 0 < x ≤ 10 | 0 < x ≤ 10 |
| Default | 1 | 1 |
Reporting bit scores
The sequence and hit reporting thresholds can also be specified as bit scores. Any sequence or hit scoring greater than or equal to that given threshold will be reported. By default, the form on the website is filled with typical values, with the sequence cut-off set to 7.0 bits and the hit cut-off set to 5.0 bits.
| Description | Sequence E-value threshold (reporting) | Hit E-value threshold (reporting) |
|---|---|---|
| Accepted Values | x > 0 | x > 0 |
| Default | 7.0 | 5.0 |
Note: These threshold scoring systems cannot be used in combination between significance and reporting, i.e. choose either significance and reporting E-values or significance and reporting bit scores when setting thresholds.
Gathering Thresholds
Gathering thresholds are defined in the respective profile HMMs. These gathering thresholds are set conservatively to ensure that there are no known false positives. Thus, if a query sequence scores with a bit score greater than or equal to the gathering thresholds, then that match is very likely to be a true positive. This threshold is the default setting for hmmscan.
Filters
Bias Composition
Turning off the bias composition filter can increases sensitivity, but at a high cost in speed, especially if the query has biased residue composition (for example a repetitive sequence region). Without the bias filter, too many sequences may pass the filter with biased queries, leading to slower than expected performance, hence by default it is switched on.
Glossary
Bit score
A bit score in HMMER is the log of the ratio of the sequence's probability according to the profile (the homology hypothesis) to the null model probability (the non-homology hypothesis).
E-value
An E-value (expectation value) is the number of hits that would be expected to have a score equal to or better than this by chance alone. A good E-value is much less than 1, for example, an E-value of 0.01 would mean that on average about 1 false positive would be expected in every 100 searches with different query sequences. An E-value around 1 is what we expect just by chance. E-values are widely used as all you need to decide on the significance of a match is the E-value, but note that they vary according to the size of the target database.
Gathering threshold
Also called the gathering cut-off, the gathering threshold is actually comprised of two bit scores, a sequence cut-off and a domain cut-off, used to define the significance of a sequence and a hit respectively. These are defined in the profile HMM and set both significance and reporting thresholds so that no insignificant hits are reported.
Profile HMM
Profile hidden Markov Models (HMMs) are a way of turning a multiple sequence alignment into a position-specific scoring system, which is suitable for searching databases for remotely homologous sequences.
Results
Identification of Cas proteins
Directly copy the 826 proteins in the Candidatus Chloracidobacterium thermophilum B provided by homepage into the input field, select E-value cut-offs, run HMMCAS and users will be shown a result page (Figure 1). Figure 1A shows the parameters users have chosen for hmmscan program. Figure 1B shows a summary table of best hits. For a query sequence matching more than one model, the model with the minimum E-value of full query sequence is treated as the best hit. The bottom left of the table shows that 23 Cas proteins are identified. If a query sequence is annotated as Cas protein, the content between the ">" character and the first space is displayed in the Query column in the table. Figure 1C shows the function that find putative cas operons from the Cas proteins which have been recognized by HMMCAS. However, there are three prerequisites for using this function:
1. Users need to be guaranteed to submit the whole proteome data;
2. Users need to ensure that the submitted proteins are ordered sequentially in the genome (Front to Back);
3. There are at least three Cas proteins that are identified by HMMCAS;
If users are sure to meet the above conditions, please click on Higher Sensitivity or Higher Specificity button to find the putative cas operon. Higher Sensitivity means an operon can tolerate up to two non-cas genes between cas genes; Higher specificity means an operon can tolerate up to one non-cas gene between cas genes. Both buttons typically result in the same outcome.
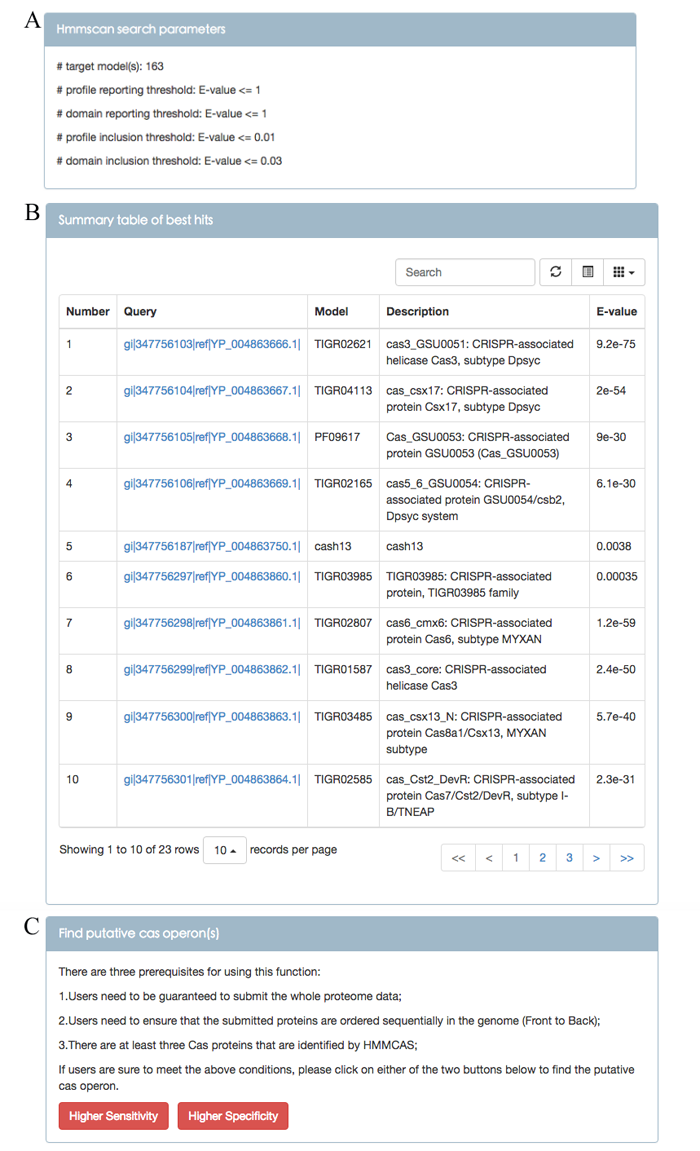
Figure 1. Screenshot of the first result page
Identification of putative cas operon
Figure 2 shows the graphical summary for cas operon architecture. Three bar graphs represent 3 different cas operons. In one cas operon, the size of Cas protein is represented by means of the number of amino acids; different Cas proteins are separated by gap that has a fixed length of 20 amino acids. From left to right and from top to bottom, the 3 different cas operons respectively consists of 4, 8 and 6 Cas proteins. When the mouse moves to the sub-bar, a message box will pop up. The message box contains the name of the Cas protein and the gene positon in the complete genome. The bottom of the Figure 2 is the scale which is represented by the number of amino acids. Users can see and compare the length of different Cas proteins via the scale.
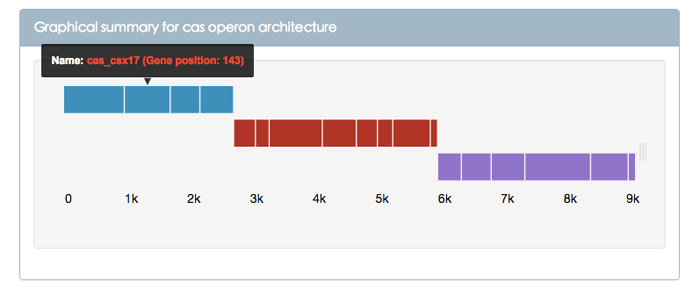
Figure 2. Graphical summary for cas operon architecture
Figure 3 is the tabular representations of cas operon found by HMMCAS. Gene position represented by positive integer means the order of the cas gene in the whole genome. In addition, the identified 3 cas operons belong to type I CRISPR-Cas system. The gene identifier with blue font in the Query column is a link to the details of corresponding Cas protein (see following Figure 4).
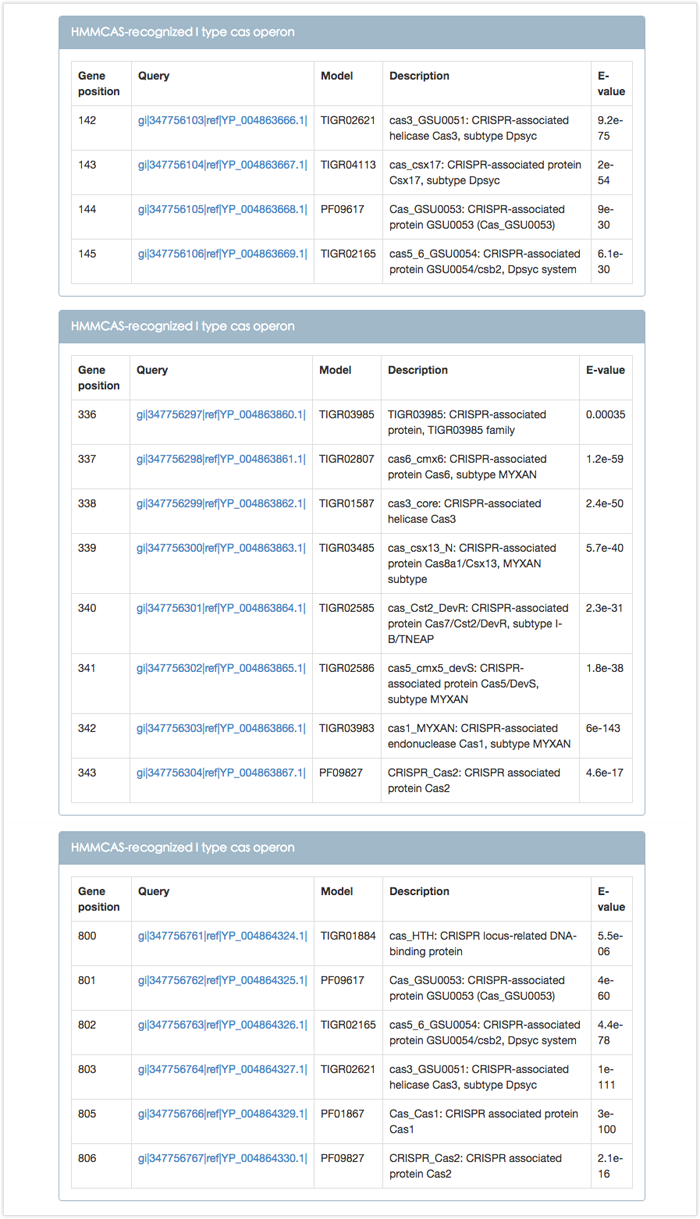
Figure 3. Tabular representations of cas operon
Details of a single Cas protein
Figure 4 shows the detailed description of a single Cas protein. Figure 4A shows the words right after the " > " symbol. The words are divided into two parts by the first space. The first part is Query line and the second part is Description line. Note that Description line is likely to be missing if there is no space in the words. Figure 4B shows the sequence information about this query. Users can click the FASTA button to download the sequence, click the CDD button to search for conserved domains within a protein, and click BLAST button to perform a sequence similarity search.
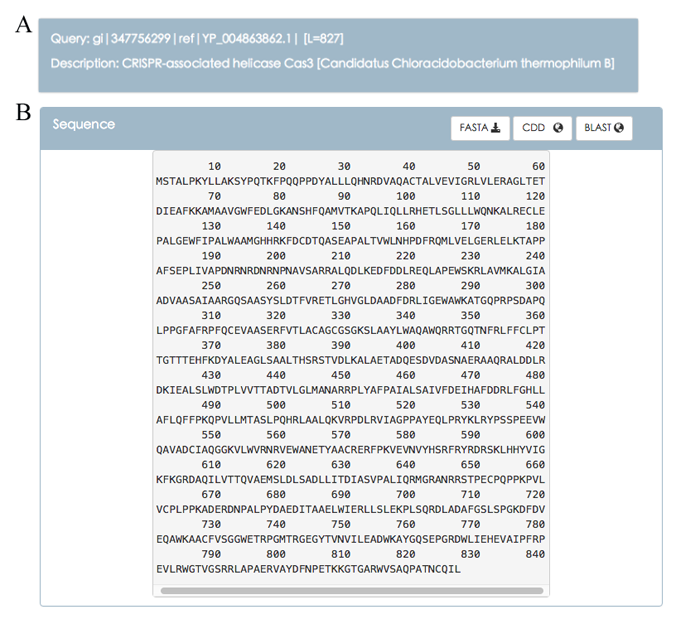
Figure 4. Description of a single Cas protein
Figure 5 shows the models matching to query shown in the Figure 4. Interval column means the subsequence of the query matching to corresponding Cas protein family HMM. As shown in Figure 5, this query is a Cas3 protein which consists of helicase and endonuclease domains. Users can click the name of model to look at the details of conserved protein domain family, and click the "+" symbol to view the detailed alignment between the query and the target model (see Figure 7, 8).
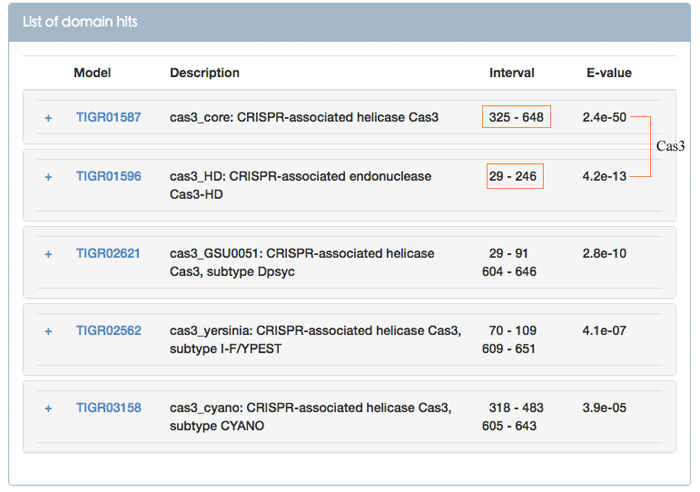
Figure 5. List of models matching to query in the Figure 4
Figure 6 shows the ability to identify the fusion protein. As shown in Figure 6, this query is a Cas1-Cas4 fusion protein.
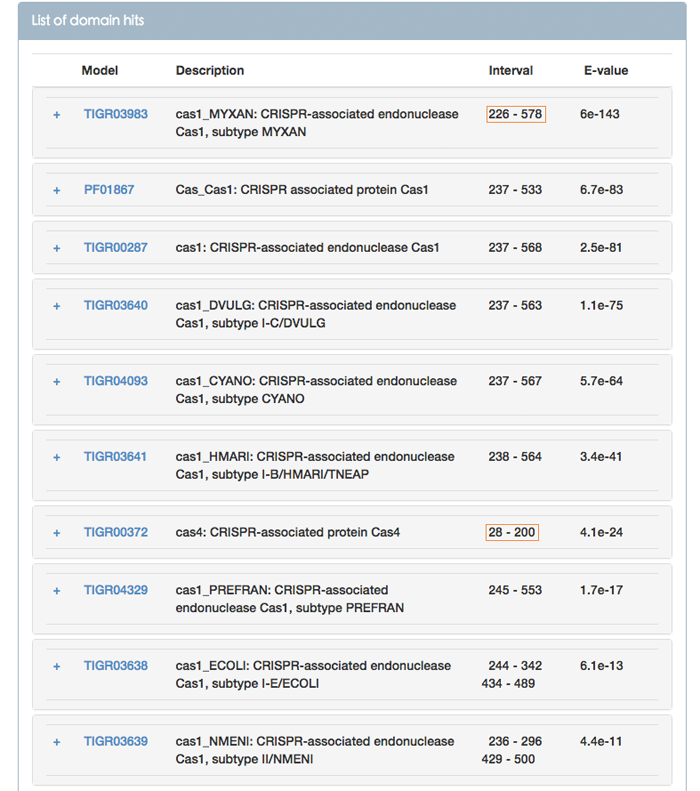
Figure 6. List of models matching to query
Alignments
Clicking on the "+" symbol displays the maximum expected accuracy (MEA) alignment between the query and the target (Figure 7). For each domain hit between the query and targets there are five rows in the alignment:
1. Header line: consists of domain number, conditional E-value and bit score for this domain.
2. Target model line: the most probable sequence from the HMM.
3. Match line: indicates identical residues (letters) or similar residues (+).
4. Query line: the sequence aligned to the model.
5. PP line: the per position posterior probability.
Bit score: the bit score for this domain.
Conditional E-value: this is the E-value that the inclusion and reporting significant thresholds that are measured against. The conditional E-value is an attempt to measure the statistical significance of each domain, given that it has already been decided that the target sequence is a true homolog. It is the expected number of additional domains or hits that would be found with a domain/hit score this big in the set of sequences reported in the top hits list, if those sequences consisted only of random non-homologous sequence outside the region that sufficed to define them as homologs.
Above the alignment the match details are presented:
Model: the name of the model. Users can click this link to look at the details of conserved protein domain family.
Description: a simple description of the model.
Interval: The start/end of the maximum expected accuracy (MEA) alignment of this domain/hit with respect to the profile HMM.
E-value: the sequence E-value that is the statistical significance of the match to this sequence. The number of hits we’d expect to score this highly in a database of this size if the database contained only non-homologous random sequences. The lower the E-value, the more significant the hit. If E-value is significant (<< 1), the sequence is likely to be a Cas protein.
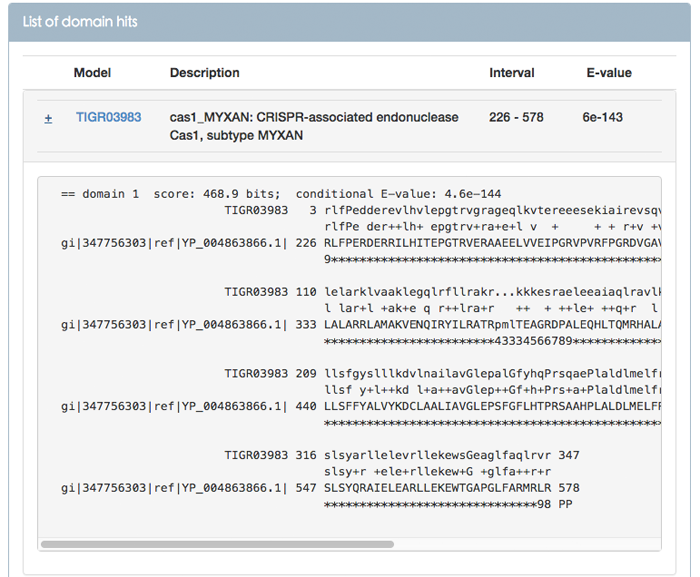
Figure 7. Alignment between the query and the target model in TIGRFAMs database
Figure 8 shows the alignment between the query and the target model in Pfam database. Compared with Figure 7, Figure 8 adds a secondary structure consensus (CS) line. This line is structural information. The following list gives the definitions for each code letter:
C: random Coil
H: alpha-helix
G: 3(10) helix
I: pi-helix
E: hydrogen bonded beta-strand (extended strand)
B: residue in isolated beta-bridge
T: h-bonded turn (3-turn, 4-turn, or 5-turn)
S: bend (five-residue bend centered at residue i)
However, not every model has the structural information.
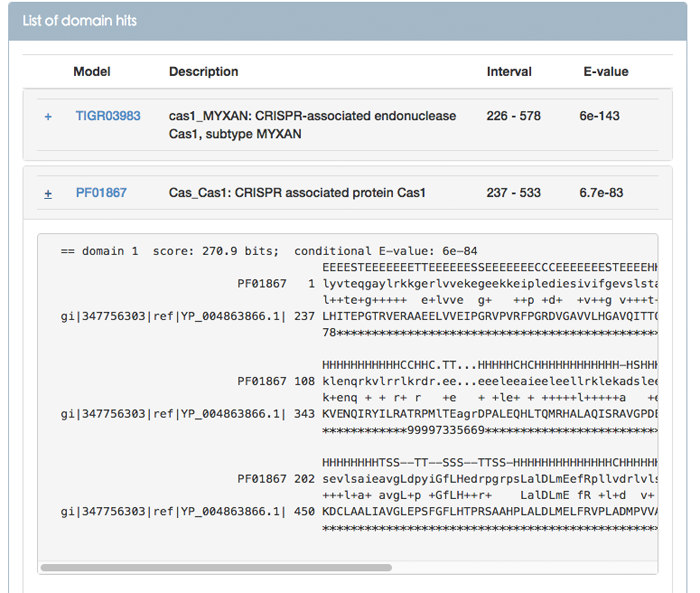
Figure 8. Alignment between the query and the target model in Pfam database
Acknowledgements
We thank the authors of Pfam and TIGRFAMs protein families databases which provide the source of Cas protein family HMMs; and Quan Zhang for continuing guidance. We thank the authors of HMMER web server due to part of our website's help page derives from this web site. We also thank the authors of HMMER software package that provides the hmmscan similarity search algorithm.