SVM, is a powerful classification method that has been widely used for protein prediction. The basic principle of SVM is to map the input vector to the high-dimensional space through the kernel function, construct the separation hyperplane with the largest spacing, and realize the classification of observations. Empirical studies have shown that the prediction performance of the radial basis kernel function (RBF) is better than the linear function, polynomial function, and sigmoid function. In this paper, we downloaded the integration toolkit LIBSVM 3.24 to implement the construction of the classification model based on SVM.
We built the final SVM model by using the training dataset (233 Ago proteins and 233 non-Ago proteins) with the 144 optimal dipeptide features, which could make full use of the dataset. The F-score of each feature was calculated based on the training dataset, and a grid search strategy was also applied on the training dataset to seek for the best feature number, the error factor c and kernel function variance gamma.
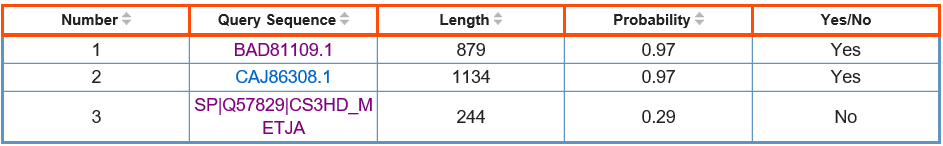
Number: the "Number" column shows the serial number of the query sequence;
Query Sequence: the "Query Sequence" column shows the content between the ">" character and the first space, with a link to the query sequence;
Length: the "Length" column shows the length of the query sequence;
Probability: the "Probability" column shows the probability that the query sequence is predicted to be a Ago protein, which is obtained based on the machine learning method of SVM;
Yes/No: the "Yes/No" column shows the prediction result, when the "Probability" is greater than or equal to "tp", the column is displayed as "Yes", otherwise "No";