MIMOX version 1.0
The first version of MIMOX was developed and published in 2006 [1]. It has two sections. In the first section, MIMOX provides a simple interface for ClustalW [2] to align a set of mimotopes. It also provides a simple statistical method to derive the motif or consensus sequence and embeds JalView [3] as a Java applet to view and manage the alignment. Surely, the embedded JalView can also be used to get a consensus sequence from the alignment of mimotopes. In the second section, MIMOX can map a single mimotope or a consensus sequence of a set of mimotopes, or parts of such sequence on to the corresponding antigen structure and search for all of the clusters of residues that could represent the native epitope. NACCESS [4] is used to evaluate the surface accessibility of the candidate clusters; and Jmol [5] is embedded to view them interactively in their 3D context.
As one of the first freely accessible web servers in this field [6], MIMOX version 1.0 is very old now. Besides, it is not automatic enough, but rather too interactive. Tuning the probe sequence fragments and parameters are often required to get good results. The interactive way gives hints to users step by step, greatly decreases the load of the server, and prevents the loss of some reasonable results. However, it only fits for experienced users with adequate background knowledge on this tool and their cases (given antibody, given antigen, and given phage display experiments). Thus, MIMOX version 1.0 was once forced to retire when MIMOX2 was ready. Perhaps, just as Dr. Dorman commented MIMOX on BioTechniques in 2006 [7]: "Clearly, computational methods would be extremely useful in order to match the mimotope with the native epitope; however, no published software has hit the sweet spot of offering flexible features while remaining platform independent and freely available. Now, Huang et al., introduce MIMOX, a web based tool for epitope mapping from phage display data...", we received a few emails from users worldwide who wrote the old version of MIMOX was still useful for them. Encouraged by the users, we reopened the faultful MIMOX version 1.0. For more help information on MIMOX version 1.0, please go to its own help page.
MIMOX2
It seems like the version 2.0 of MIMOX. However, their algorithms and design philosophies are quite different. Unlike MIMOX version 1.0, MIMOX2 is more automatic and friendly. Users only need to provide the antigen structure (PDB file) and the mimotope sequences and then click the decode button. At first, MIMOX2 will divide the antigen surface into overlapping patches. Then it will evaluate each surface patch's similarity to each mimotope according to the compositions and physicochemical properties of their residues. The surface patches which are similar to the set of mimotopes are combined and predicted as epitopes. There are two ways to provide the PDB file: uploading the PDB file or just typing in the PDB ID and the chain name. Please remove all unnecessary chains and only keep the antigen chain when you upload a PDB file.
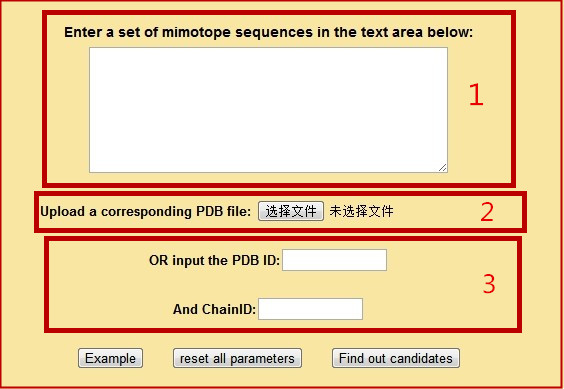
Look at the interface below. The region 1 is used for entering mimotope sequences. The region 2 is used for uploading the PDB file, the region 3 is used for typing in the PDB ID and the chain name.
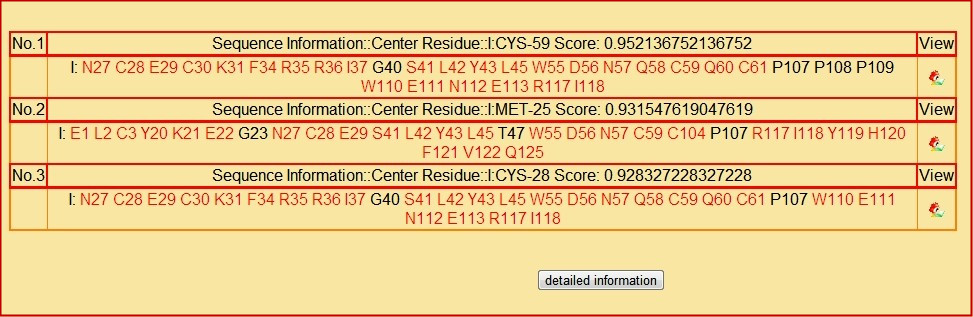
After the analysis, as the picture shown below, the top 3 patches which have the highes score will be shown. You can see the amino acids contained in the patches, among which the amino acids that have a high possiblity to be epitopes will be shown in red. Users can also get the patch information such as the patch center and the patch score in the result. The 'view' column is the entrance to the 3D view of the corresponding structure.
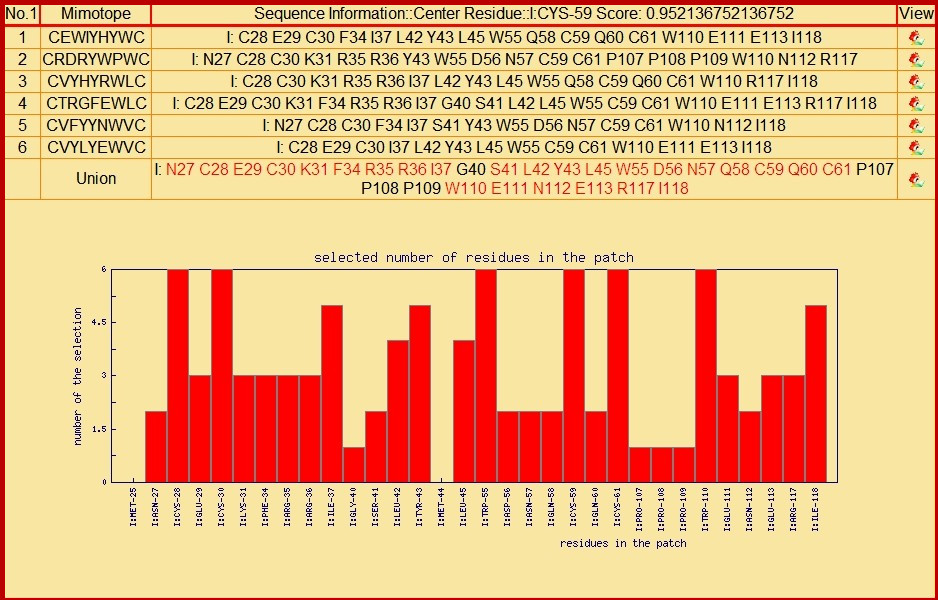
If user push the 'detailed information' button, all patches and its corresponding detailed information will be shown in accordance with the order of score. See below, it is the information of No.1 patch. In it, users can see amino acids similar to each mimotope. The 'Union' is a sum of amino acids appeared above, the amino acids appeared twice and more are drawn in red. The Histogram, as a reference, show the occurrence number of each amino acids.
PyMIMOX
PyMIMOX is a PyMOL plugin of MIMOX which is under developemnt currently...
References
- Huang J, Gutteridge A, Honda W, Kanehisa M. MIMOX: a web tool for phage display based epitope mapping. BMC Bioinformatics. 2006; 7: 451.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680.
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427.
- Hubbard SJ, Thornton JM. NACCESS. Department of Biochemistry and Molecular Biology, University College London; 1993.
- Jmol http://jmol.sourceforge.net
- Knittelfelder R, Riemer AB, Jensen-Jarolim E. Mimotope vaccination--from allergy to cancer. Expert Opin Biol Ther. 2009; 9(4): 493-506.
- Dorman N. A Perfect Fit. Biotechniques. 2006; 41(6): 655.