Help Contents:
The AGO3D Search Algorithm: BLASTP
The BLASTP is used to search protein sequences against proteins of built database with MAKEBLASTDB algorithm. AGO3D uses BLAST similarity search algorithm in version 2.2.31 which downloads from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ to search against the proteins containing PIWI domain and MID domain.
The AGO3D Modeling Algorithm: MODELLER
The MODELLER is used to construct three-dimensional structure of Ago protein. AGO3D uses MODELLER in version 19.9 which download from https://salilab.org/modeller/download_installation.html to select the best structure template and then construct the modeling.
Input Data
Only one-letter codes for 20 common amino acids and X letter are accepted, and any other characters will lead to an illegal character feedback. The input can be only one protein sequence and the formation of protein is ali formation.We have provided an example file with NgAgo protein sequence. Figure 1 shows the input field and the ali formation.
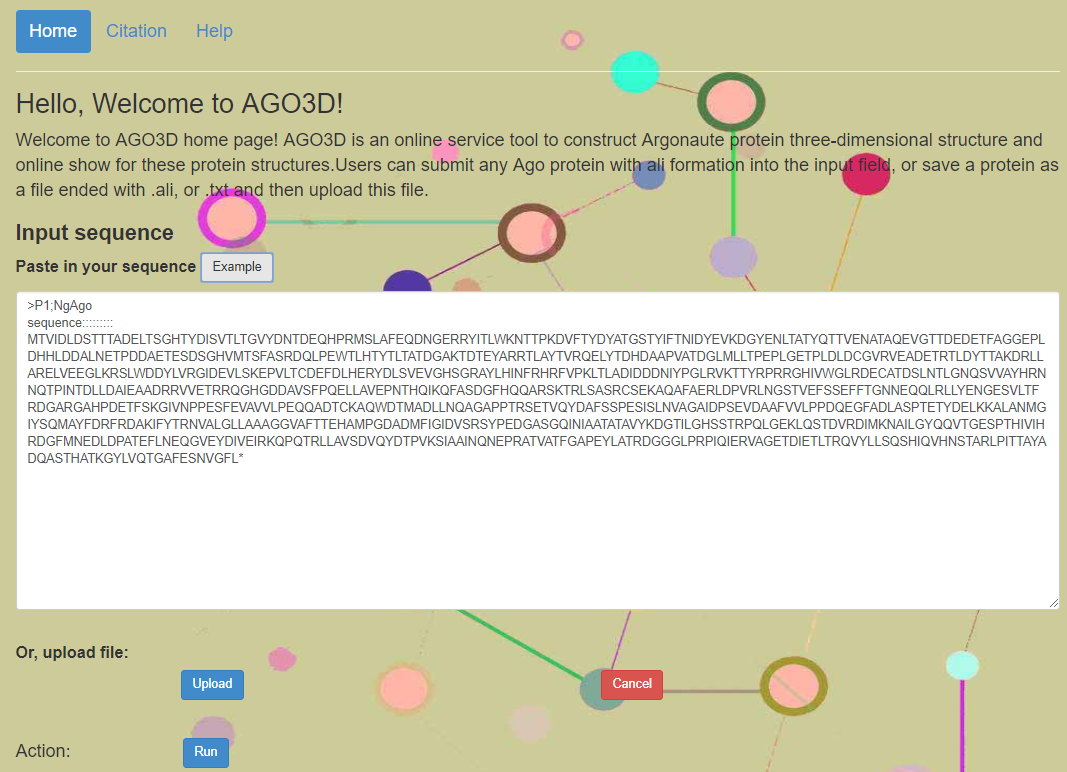
Figure 1. Screenshot of the input page
Results
Directly click the example button, one example provided by homepage into the input field, run AGO3D and users will obtain a waitting page (Figure 2). Figure2 shows that one mouth is eating eating amino acid residue one by one. And it also displays the running time in seconds. When the models are built, users can get a result page (Figure 3). Figure 3 has a three-dimensional structure display window in JSmol. There are five model buttons under JSmol window to choose to display. And the bottom of this figure also shows download button of the corresponding five model files ended .pdb.
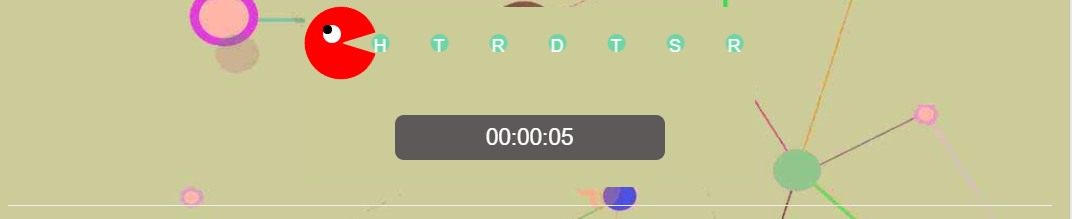
Figure 2. Waitting page
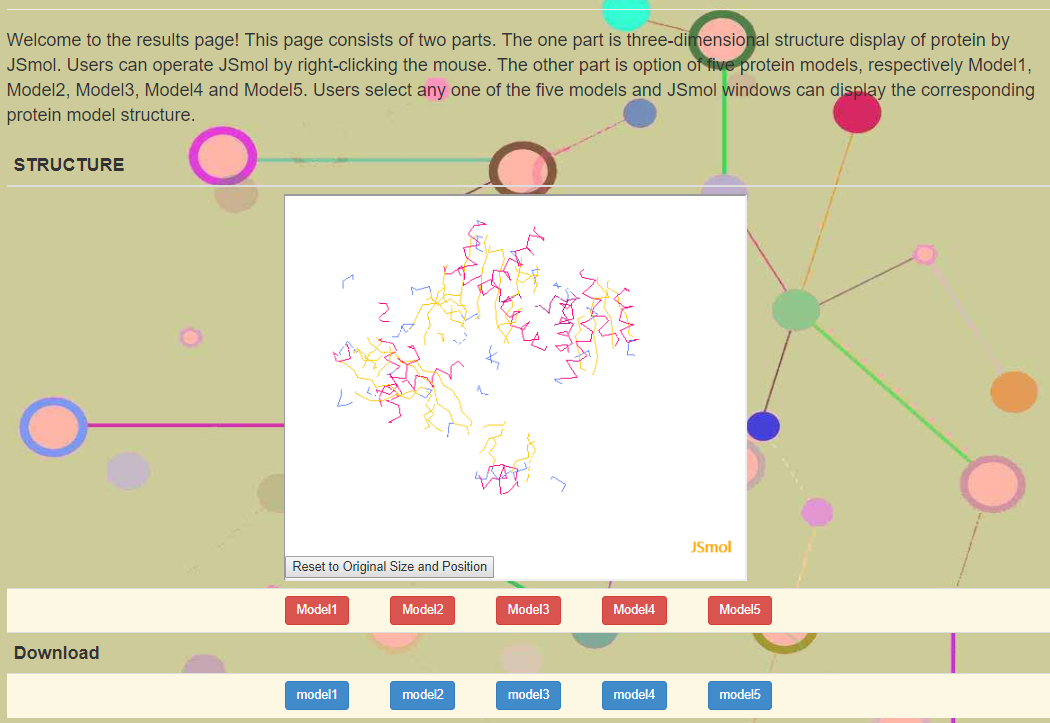
Figure 3. Results page