Help
Overview:
The Home page of AACDB Version 1.0 provides a detailed description of the database's functionalities. This comprehensive database comprises 7498 antigen-antibody complex entries, encompassing antibodies from over 14 different organisms and detailed classification into 16 antibody types. It offers users a user-friendly interface to effortlessly query, manipulate, browse, and visualize the sequences, structures, and distances of antibody-antigen complexes. This facilitates the identification of paratopes and epitopes, enabling a deeper understanding of the complex interactions.
The Datasets page is displayed in Fig. 1:
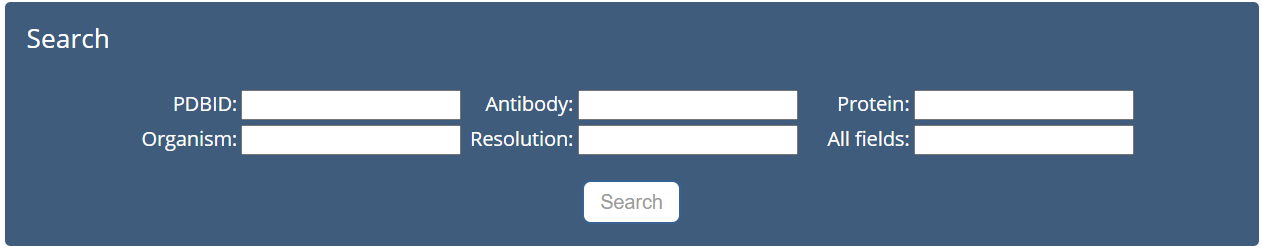
Fig. 1. Search page
On Datasets page, we can enter keywords to search the sequence and structural information about antibody/antigen/antibody-antigen complex. The AACDB database supports fuzzy search and multi-field search.
Search through the following processes:
(1) Select one or more keyword types as input:
We provide a total of 6 keyword types: PDBID, Antibody, Protein, Organism, Resolution and All fields.
PDBID: The unique identifier of the RCSB PDB database (https://www.rcsb.org/).
Antibody: The user can provide name/ aliase/ fragment of an antibody, change the keywords to finally get the information you want.
Protein: In our data, antigens only collect macromolecular proteins, and by searching the name of the protein/ antigen you can get the results you want.
Organism: Users can obtain information about antibody-antigen complexes by searching the organism source of antibodies or antigens.
Resolution: Resolution obtained by X-ray diffraction (XRD), electron microscopy (EM) or nuclear magnetic resonance (NMR) in the RSCB PDB structure database. Users can filter by customizing the cut off value of the resolution (Å).
All fields: Users can use this function to search the entire database, regardless of the keyword type.
(2) click Search
(3) understanding of result:
The following is a simple example using keyword type = Antibody, keyword = Fab. Figure 2 shows the result table of the top 10 antibody-antigen complexes of the Fab antibody.
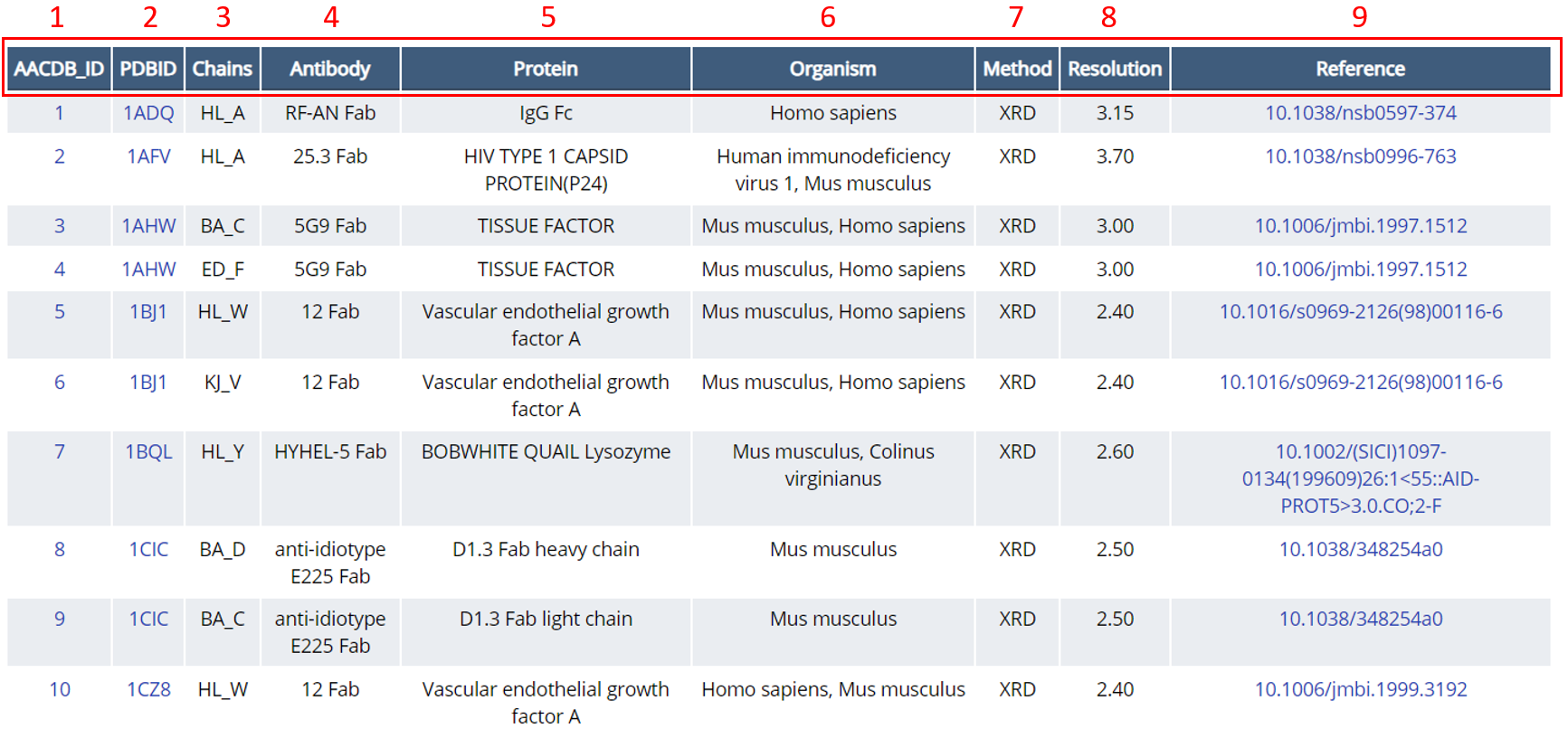
Fig. 2 Top ten entries of antibody-antigen complexes indexed using "Antibody = Fab" as keyword
1 ACCDB_ID: The unique identifier of the AACDB. Clicking on each ACCDB_ID will turn to the Details page of this entry, the interpretation of which will be explained later.
2 PDBID: The unique identifier of the RCSB PDB database. Clicking on each PDBID will turn to the entry in RCSB PDB database.
3 Chain: The entry in this column is the antibody-antigen complex chain after manual calibration, which is presented in the format of “antibody heavy chain antibody light chain_antigen chain” (e.g. HL_A).
4 Antibody: Antibody name + antibody fragment (e.g.HyHEL-63 Fab/NC10 Fv/Nb5776 nanobody...).
5 Protein: Large molecule antigen proteins (length of aa >50).
6 Organism:The organism source of antibodies or antigens.
7 Method: Three methods for observing the structure of antibody-antigen complex: X-ray diffraction (XRD), electron microscopy (EM) and nuclear magnetic resonance (NMR).
8 Resolution: The resolution (Å) corresponding to the above three methods.
9 Reference: Each antibody-antigen complex has been linked to the original published literature.
Users can finally download the search results according to the keywords to "txt" or "csv" result file. It can also download all 7498 entries to "txt" or "csv" result file by clicking 1 and 2 as following in Fig. 3 respectively. Users can use this function to achieve accurate or batch downloads.

Fig. 3 Download the result in Datasets page
To view of details of the entry in AACDB, you should click the “AACDB_ID” at the front-end of Datasets page.
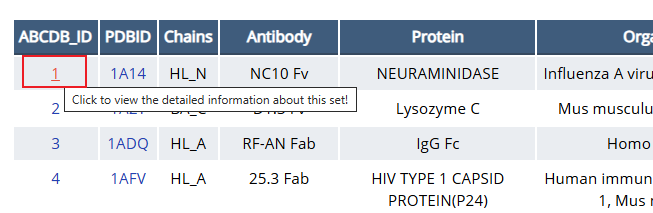
Fig. 4 Click the “AACDB_ID” to view the detailed information
(1) Structure information
In the Details page, you can get the detail information of the antibody-antigen complex. Take “PDBID = 1ADQ” as an example. Click 1 in the red box in Fig. 4 to turn to the Details page of 1ADQ as shown in Fig. 5.
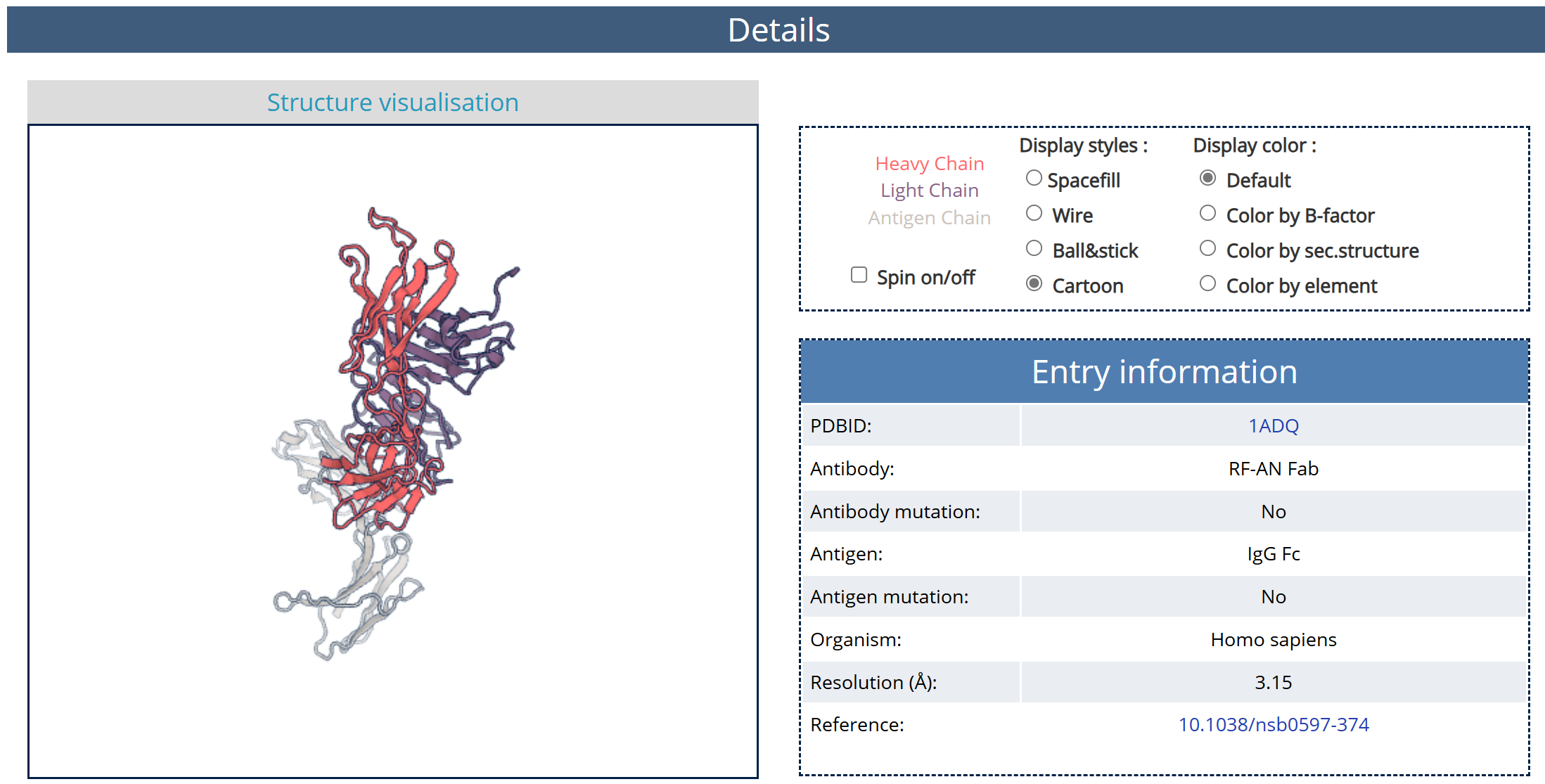
Fig. 5 structure visualization of 1ADQ
In this part we provied visualizaton of the structure of the antibody-antigen complex 1ADQ with optional different display styles and colors. The antibody and antigen of this complex are listed in the Entry information.
(2) Sequence information
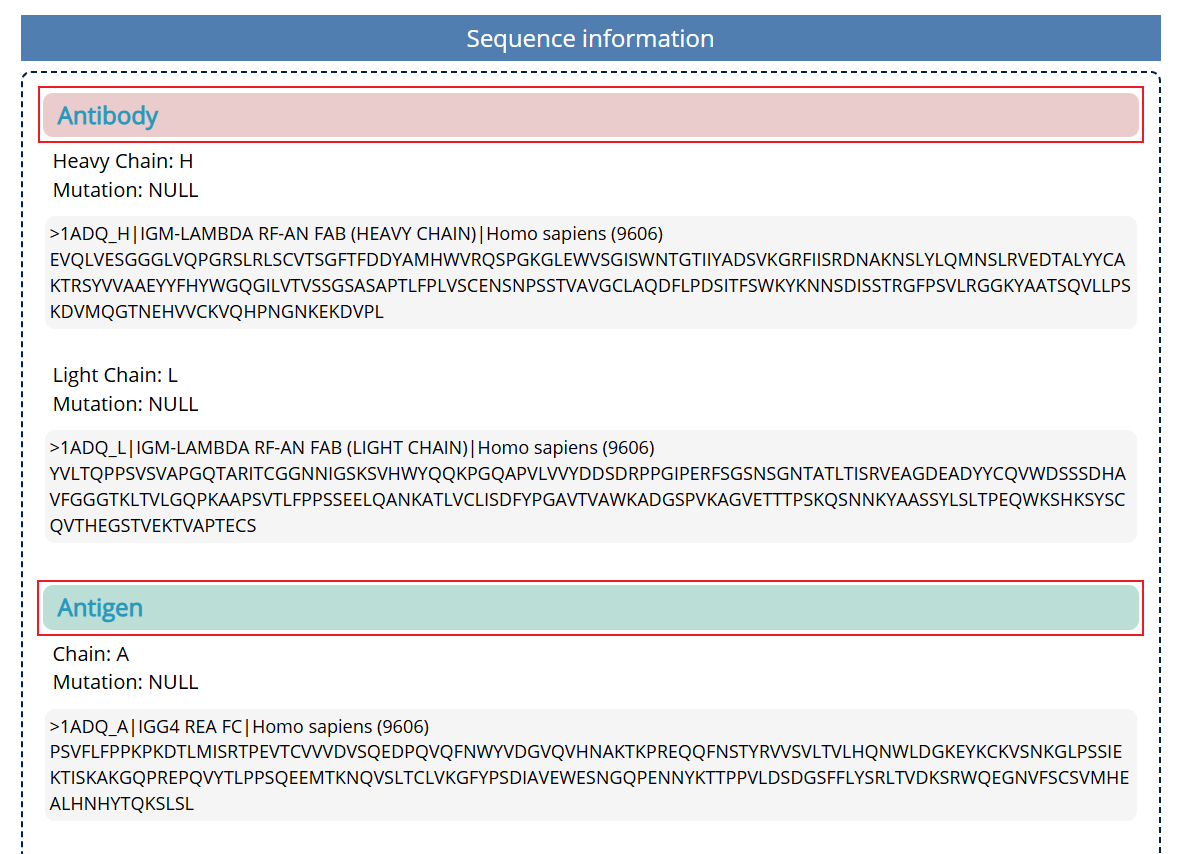
Fig. 6 The heavy/ light chain of antibody and antigen in 1ADQ
As shown in Fig. 6, in this part, you can browse FASTA sequence and mutation information for the heavy/ light chain of antibody and antigen.
(3) Interaction information

Fig. 7 Detailed information on interacting residues
As shown in Fig. 7, AACDB provides detailed information on interacting residues using two methods (ΔSASA and atom distance), facilitating the identification of paratopes and epitopes.
(4) Download
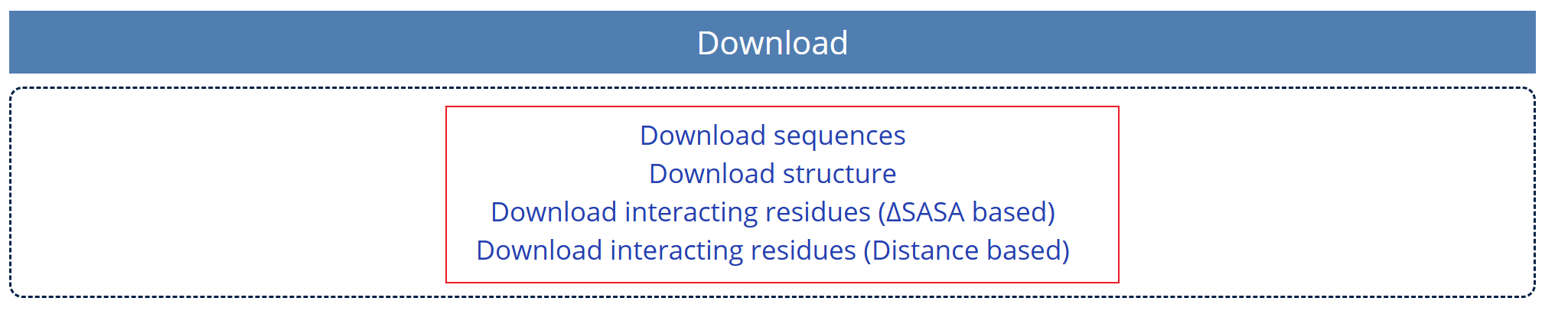
Fig. 8 Download in the Details Page
The four datas shown in Fig. 8 can be downloaded from this part.
We provide two ways for downloading the data:
(1) Download data of the single entry.
When access the detailed information about an entry using the corresponding AACDB_ID, user can click the linker at the “Download” section of the bottom of the page to download the data for a single certain entry.

Fig. 9 Download in the Details Page
(2) Download all the data of AACDB.
AACDB provide the download page for users. You can download all the sequence, structure and interacting data in the download page. The sequence and structure file of antibody, antigen and complex were packaged in different .zip file, respectively. The interacting data based on different method were packaged in different .zip file too.
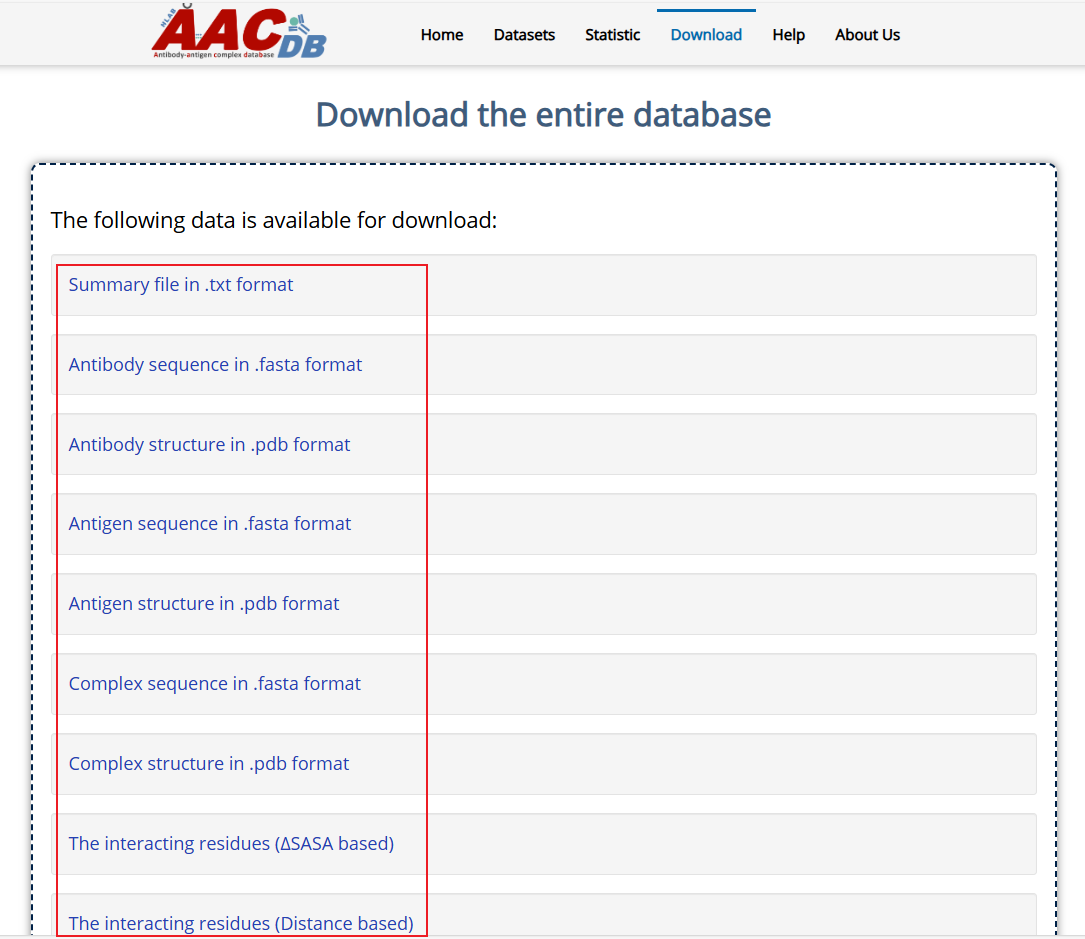
Fig. 10 Download in the Download Page
All the entries in the database were manually searched and verified according to the RCSB PDB database entries and the original literature, and we uniformed the description of the complex chains. We carefully checked each entry manually to find out all possible annotation problems and did find a lot of annotation absence and errors. In the AACDB, we have supplemented the available information or correct the annotations through comprehensive literature reviews and sequence alignment with wild-type proteins.
Antibody complexes in the RCSB PDB database, up to December 2023, were downloaded. At present, the AACDB database is focusing on the complexes between antibodies and protein antigens. Therefore, we filtered out antibody-peptide (< 50 aa) complexes, antibody-nucleic acid complexes and antibody-small molecule complexes. We also excluded the antigen that bind to Fc fragment of antibody and the crystallization partners (generally nanobody/ VHH) that assist macromolecular protein crystallization.
The RCSB PDB database provides the two PDB chain IDs (label_asym_id; assigned by the PDB) and auth_asym_id (selected by the author). We usually chose the auth_asym_id chain to represent the chain id of the antibody/antigen. In some special cases, the length of auth_asym_id longer than one, we use the simple label_asym_id as chain id. For example, in entry 7B27, the chains annotation of “Surface glycoprotein” is “A [auth AAA]”, we use the “A” as chain id in AACDB rather than “AAA”.
The antibody nomenclature follows the title of the corresponding search entry in the RCSB PDB database, with verification done through the original literature. In cases where the name in the original literature differs from that in the RCSB PDB, we used the name in the published literature as the standard. Furthermore, for antibody fragments lacking names in both the RCSB PDB database and original literature, we adopt a naming convention of "PDBID" + "antibody fragment" (e.g., 4WEB Fab).
The PDB files containing multiple antibodies were split into separate files, each containing antigen chains as appropriate. For example, PDB file 1DZB contains 2 copies of the same antibody and antigen, it was split into 2 entries that including A_X and B_Y chains respectively.
(1) View of data: If you directly open those downloaded txt or csv format files using Office Excel, you may encounter certain issues. For example, the PDBID "4JAN" might be mistakenly recognized as a date and displayed as "4-Jan". Similarly, the value "7E23" can be misinterpreted as a number and displayed as "7.00E+23" due to the Excel version or settings. To avoid these problems, we recommend following the steps outlined in Figure 11 or opening the files with a text editor such as Notepad++.
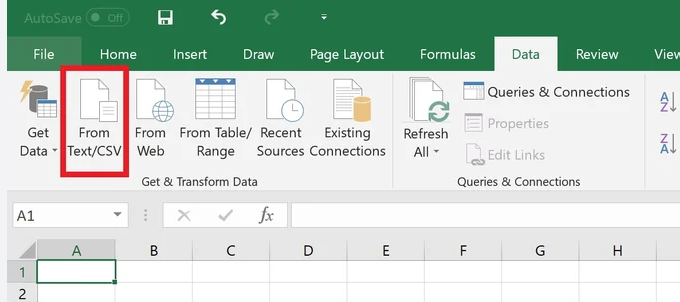
Fig. 11 Steps of data view in Excel
(2) Limitations:Although we have made our best effort, the limitations of our team's resources and scope of knowledge mean that our database may not capture all antigen-antibody complex structures. If you come across any missed structures in our database, we kindly hope that you contact us and provide detailed information to ensure accurate data integration. We greatly appreciate your valuable contributions to AACDB.
(3) Shortcomings: As the data is manually collected, it is challenging to completely eliminate errors during the information processing. If you come across any inaccuracies in the current database, we kindly request that you promptly communicate these issues to us via email. Your feedback will greatly assist us in improving the accuracy of our data.
(1) Load the antigen-antibody complex crystal file and named as "complex"
PyMOL>load ./aacdb/1A14_HLN.pdb, complex
(2) Color the complex by chains
PyMOL>util.cbc("complex")
(3) Select antibody and antigen chains
PyMOL>select antibody, chain H or chain L
PyMOL>select antigen, chain N
(4) Select interface (To identify the interaction interface, residues within a specified distance threshold (default: 6 Å) between the antibody and antigen were selected. This distance can be adjusted based on the specific requirements of your analysis.)
PyMOL>select interface, (antibody within 6 of antigen) or (antigen within 6 of antibody)
(5) Highlight the interface
PyMOL>show sticks, interface
PyMOL>color tv_blue, antibody and interface
PyMOL>color tv_blue, antigen and interface
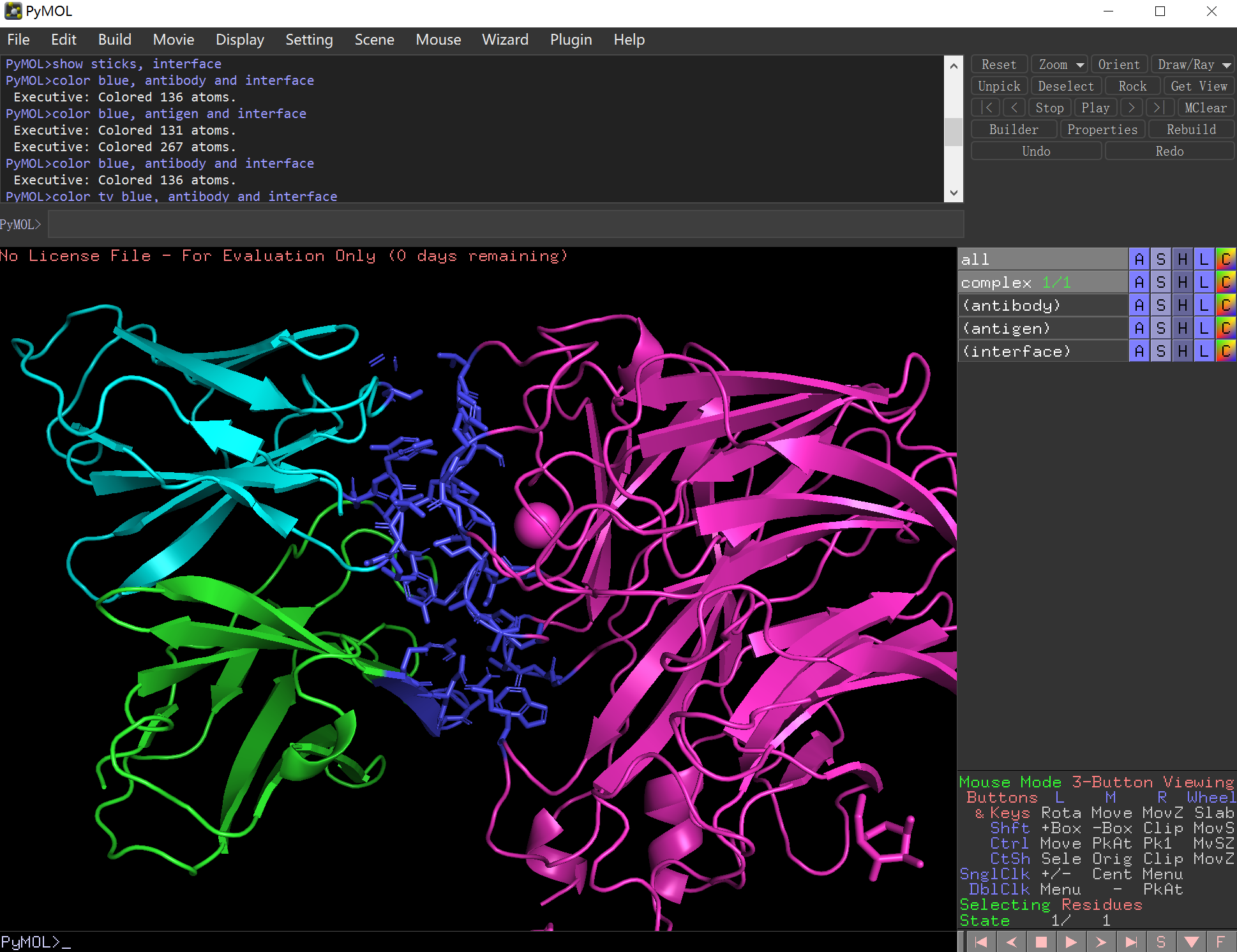
Fig. 12 Interface definition in pymol