Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 856 |
| PDBID: | 4NZR |
| Chains: | HL_M |
| Organism: | Homo sapiens, Mycoplasmoides genitalium G37 |
| Method: | XRD |
| Resolution (Å): | 1.65 |
| Reference: | 10.1126/science.1246135 |
| Antibody | |
| Antibody: | PGT135 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | Protein M TD |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Heavy Chain: H
Mutation: NULL
| >4NZR_H|Chain A[auth H]|PGT135 heavy chain|Homo sapiens (9606) QLQMQESGPGLVKPSETLSLSCTVSGDSIRGGEWGDKDYHWGWVRHSAGKGLEWIGSIHWRGTTHYKESLRRRVSMSIDTSRNWFSLRLASVTAADTAVYFCARHRHHDVFMLVPIAGWFDVWGPGVQVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKRVEPK |
Light Chain: L
Mutation: NULL
| >4NZR_L|Chain B[auth L]|PGT135 light chain|Homo sapiens (9606) EIVMTQSPDTLSVSPGETVTLSCRASQNINKNLAWYQYKPGQSPRLVIFETYSKIAAFPARFVASGSGTEFTLTINNMQSEDVAVYYCQQYEEWPRTFGQGTKVDIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC |
Antigen
Chain: M
Mutation: NULL
| >4NZR_M|Chain C[auth M]|Protein M TD|Mycoplasma genitalium (243273) MGSSHHHHHHSSGLVPRGSHMSLSLNDGSYQSEIDLSGGANFREKFRNFANELSEAITNSPKGLDRPVPKTEISGLIKTGDNFITPSFKAGYYDHVASDGSLLSYYQSTEYFNNRVLMPILQTTNGTLMANNRGYDDVFRQVPSFSGWSNTKATTVSTSNNLTYDKWTYFAAKGSPLYDSYPNHFFEDVKTLAIDAKDISALKTTIDSEKPTYLIIRGLSGNGSQLNELQLPESVKKVSLYGDYTGVNVAKQIFANVVELEFYSTSKANSFGFNPLVLGSKTNVIYDLFASKPFTHIDLTQVTLQNSDNSAIDANKLKQAVGDIYNYRRFERQFQGYFAGGYIDKYLVKNVNTNKDSDDDLVYRSLKELNLHLEEAYREGDNTYYRVNENYYPGASIYENERASRDSEFQNEILKR |
Interaction
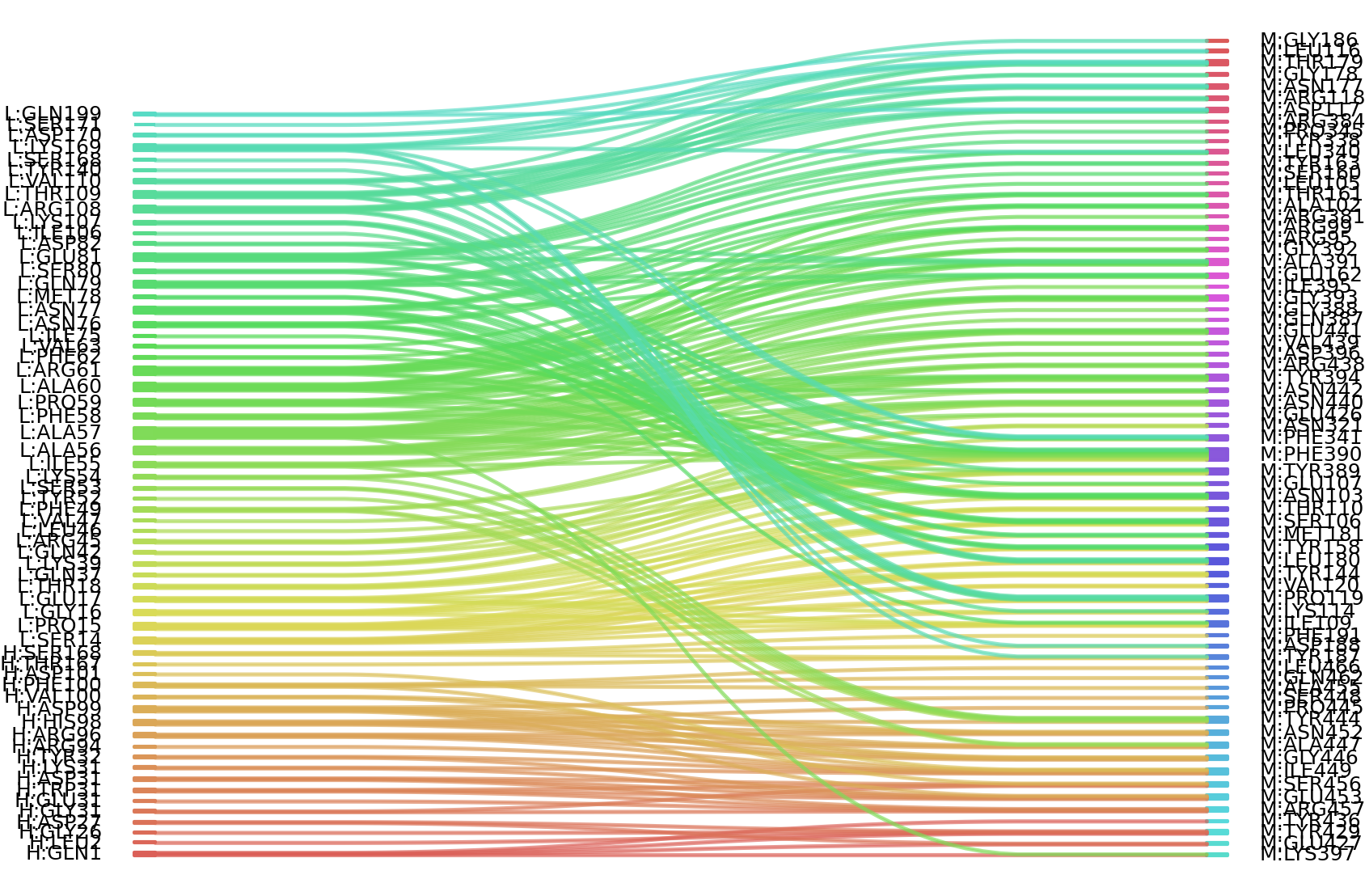
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition H: GLN1 GLY26 ASP27 TYR32 ARG94 ARG96 HIS98 ASP99 THR167 SER168 GLY169 L: SER14 PRO15 GLY16 GLU17 THR18 GLN37 LYS39 GLN42 ARG45 VAL47 PHE49 TYR52 SER53 LYS54 ILE55 ALA56 ALA57 PHE58 PRO59 ALA60 ARG61 PHE62 VAL63 ASN76 ASN77 GLN79 SER80 GLU81 ASP82 LYS107 ARG108 THR109 VAL110 SER168 LYS169 ASP170 SER171 GLN199 M: ARG95 ARG99 ALA102 ASN103 SER106 GLU107 ILE109 THR110 LYS114 LEU116 ASP117 ARG118 PRO119 TYR144 TYR158 SER160 THR161 GLU162 TYR163 ASN177 GLY178 THR179 LEU180 MET181 GLY186 TYR187 ASP188 PHE191 ASN321 TYR338 LEU340 PHE341 ARG384 TYR389 PHE390 ALA391 GLY392 GLY393 TYR394 ASP396 LYS397 GLU426 GLU427 TYR429 TYR436 ARG438 VAL439 ASN440 GLU441 ASN442 TYR444 PRO445 GLY446 ALA447 ILE449 ASN452 GLU453 SER456 ARG457 GLN462 LEU466 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)