Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 7436 |
| PDBID: | 7ANQ |
| Chains: | B_A |
| Organism: | Homo sapiens, Lama glama |
| Method: | XRD |
| Resolution (Å): | 2.20 |
| Reference: | 10.1016/j.molmet.2022.101662 |
| Antibody | |
| Antibody: | P1.40 VHH |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | Proprotein convertase subtilisin/kexin type 9 (PCSK9) |
| Antigen mutation: | No |
| Durg Target: | Q8NBP7 |
Sequence information
Antibody
Chain: B
Mutation: NULL
| >7ANQ_B|Chain B|VHH P1.40 minibody anti-Cter PCSK9|Lama glama (9844) QVKLEESGGGLVQAGGSLRLSCSPSDRTFSAYAMGWFRQVPGREREFVATIRDSDASIYYTDSVKGRFTISRDNAKNTVYLQMNSLIPDDTAVYYCAARQYYSGRVYSTFREEYDYWGQGTQVTVSS |
Antigen
Chain: A
Mutation: NULL
| >7ANQ_A|Chain A|Proprotein convertase subtilisin/kexin type 9|Homo sapiens (9606) GWQLFCRTVWSAHSGPTRMATAVARCAPDEELLSCSSFSRSGKRRGERMEAQGGKLVCRAHNAFGGEGVYAIARCCLLPQANCSVHTAPPAEASMGTRVHCHQQGHVLTGCSSHWEVEDLGTHKPPVLRPRGQPNQCVGHREASIHASCCHAPGLECKVKEHGIPAPQEQVTVACEEGWTLTGCSALPGTSHVLGAYAVDNTCVVRSRDVSTTGSTSEGAVTAVAICCRSR |
Interaction
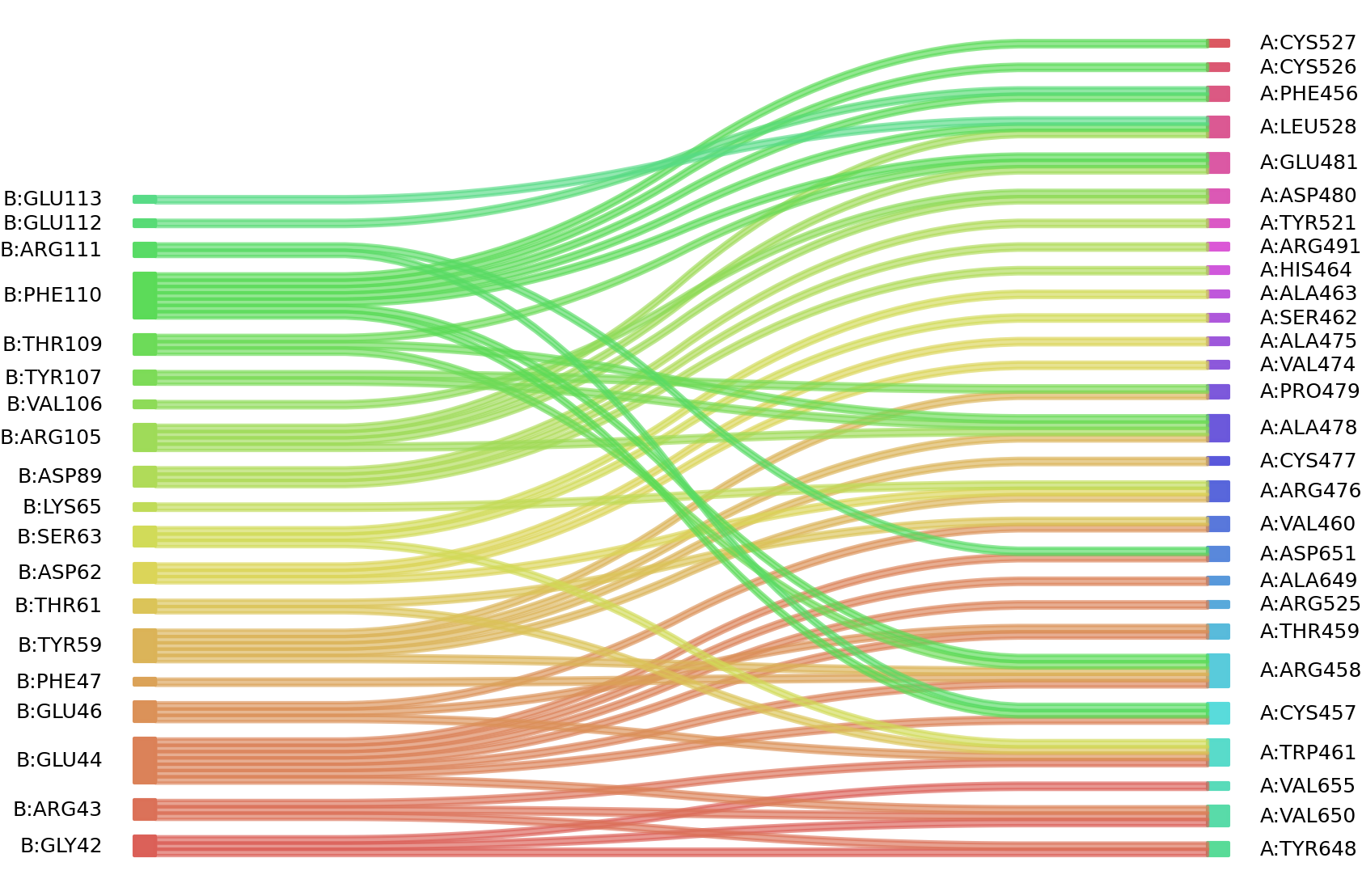
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition B: GLY42 ARG43 GLU44 GLU46 PHE47 TYR59 THR61 ASP62 SER63 LYS65 ILE87 ASP89 ARG105 VAL106 TYR107 THR109 PHE110 ARG111 GLU112 GLU113 A: PHE456 CYS457 ARG458 THR459 VAL460 TRP461 SER462 ALA463 HIS464 VAL474 ARG476 CYS477 ALA478 PRO479 ASP480 GLU481 ARG491 TYR521 ARG525 CYS526 CYS527 LEU528 TYR648 ALA649 VAL650 ASP651 VAL655 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)