Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 6572 |
| PDBID: | 1BGX |
| Chains: | HL_T |
| Organism: | Thermus aquaticus, Mus musculus |
| Method: | XRD |
| Resolution (Å): | 2.30 |
| Reference: | 10.1073/pnas.95.21.12562 |
| Antibody | |
| Antibody: | TP7 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | Thermus aquaticus DNA polymerase |
| Antigen mutation: | No |
| Durg Target: | P19821 |
Sequence information
Antibody
Heavy Chain: H
Mutation: NULL
| >1BGX_H|Chain C[auth H]|TP7 MAB|Mus musculus (10090) EVQLQESGPGLVKPYQSLSLSCTVTGYSITSDYAWNWIRQFPGNKLEWMGYITYSGTTDYNPSLKSRISITRDTSKNQFFLQLNSVTTEDTATYYCARYYYGYWYFDVWGQGTTLTVSSAKTTAPSVYPLAPVSSVTLGCLVKGYFPEPVTLTWNSGSLSSGVHTFPAVLQSDLYTLSSSVTVTSSTWPSQSITCNVAHPASSTKVDKKIEPR |
Light Chain: L
Mutation: NULL
| >1BGX_L|Chain B[auth L]|TP7 MAB|Mus musculus (10090) DIQMTQSPAIMSASPGEKVTMTCSASSSVSYMYWYQQKPGSSPRLLIYDSTNLASGVPVRFSGSGSGTSYSLTISRMEAEDAATYYCQQWSTYPLTFGAGTKLELKRADAAPTVSIFPPSSEQLTSGGASVVCFLNNFYPKDINVKWKIDGSERQNGVLNSWTDQDSKDSTYSMSSTLTLTKDEYERHNSYTCEATHKTSTSPIVKSFNR |
Antigen
Chain: T
Mutation: NULL
| >1BGX_T|Chain A[auth T]|TAQ DNA POLYMERASE|Thermus aquaticus (271) MRGMLPLFEPKGRVLLVDGHHLAYRTFHALKGLTTSRGEPVQAVYGFAKSLLKALKEDGDAVIVVFDAKAPSFRHEAYGGYKAGRAPTPEDFPRQLALIKELVDLLGLARLEVPGYEADDVLASLAKKAEKEGYEVRILTADKDLYQLLSDRIHVLHPEGYLITPAWLWEKYGLRPDQWADYRALTGDESDNLPGVKGIGEKTARKLLEEWGSLEALLKNLDRLKPAIREKILAHMDDLKLSWDLAKVRTDLPLEVDFAKRREPDRERLRAFLERLEFGSLLHEFGLLESPKALEEAPWPPPEGAFVGFVLSRKEPMWADLLALAAARGGRVHRAPEPYKALRDLKEARGLLAKDLSVLALREGLGLPPGDDPMLLAYLLDPSNTTPEGVARRYGGEWTEEAGERAALSERLFANLWGRLEGEERLLWLYREVERPLSAVLAHMEATGVRLDVAYLRALSLEVAEEIARLEAEVFRLAGHPFNLNSRDQLERVLFDELGLPAIGKTEKTGKRSTSAAVLEALREAHPIVEKILQYRELTKLKSTYIDPLPDLIHPRTGRLHTRFNQTATATGRLSSSDPNLQNIPVRTPLGQRIRRAFIAEEGWLLVALDYSQIELRVLAHLSGDENLIRVFQEGRDIHTETASWMFGVPREAVDPLMRRAAKTINFGVLYGMSAHRLSQELAIPYEEAQAFIERYFQSFPKVRAWIEKTLEEGRRRGYVETLFGRRRYVPDLEARVKSVREAAERMAFNMPVQGTAADLMKLAMVKLFPRLEEMGARMLLQVHDELVLEAPKERAEAVARLAKEVMEGVYPLAVPLEVEVGIGEDWLSAKE |
Interaction
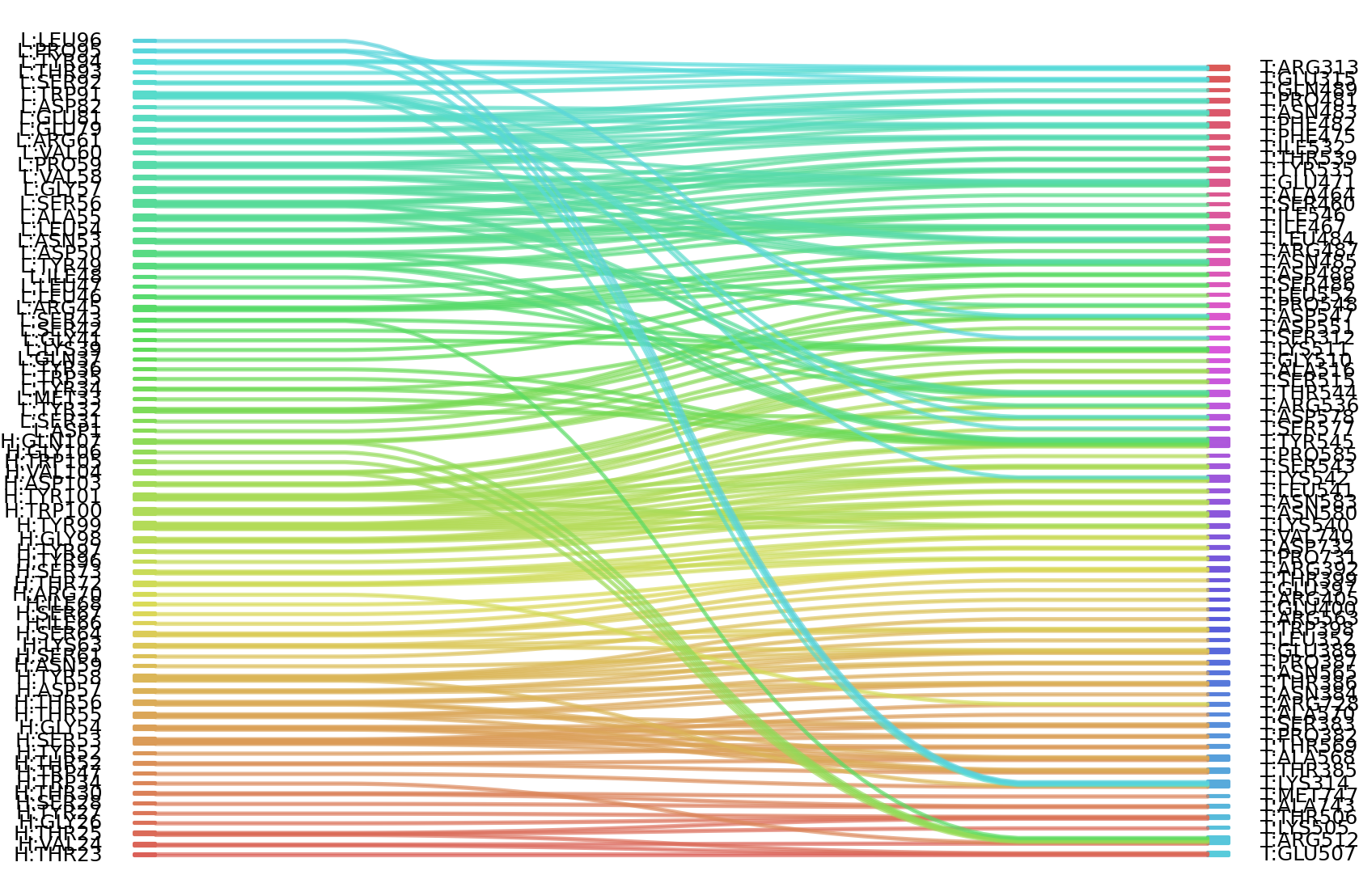
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition L: ASP1 SER31 TYR32 TYR34 TRP35 TYR36 GLN37 LYS39 GLY41 SER42 SER43 ARG45 LEU46 LEU47 ILE48 TYR49 ASP50 SER51 ASN53 LEU54 ALA55 SER56 GLY57 VAL58 PRO59 VAL60 ARG61 PHE62 SER67 GLU79 GLU81 ASP82 TRP91 SER92 THR93 TYR94 PRO95 LEU96 SER168 H: GLN5 THR23 VAL24 THR25 GLY26 TYR27 SER28 THR30 ASP31 TRP34 TRP47 TYR50 TYR52 SER53 GLY54 THR55 THR56 ASP57 TYR58 ASN59 SER61 LEU62 LYS63 SER64 ILE66 SER67 ILE68 THR69 ARG70 THR72 SER73 ASN75 CYS92 ARG94 TYR96 TYR97 GLY98 TYR99 TRP100 TYR101 PHE102 ASP103 VAL104 TRP105 GLY106 GLN107 T: TRP167 PHE309 LEU311 SER312 ARG313 LYS314 GLU315 LEU352 PRO382 SER383 THR385 THR386 PRO387 GLU388 GLY389 ARG392 GLU397 TRP398 THR399 GLU400 ARG405 SER460 ALA464 ILE467 ALA468 GLU471 ALA472 PHE475 PRO481 PHE482 ASN483 LEU484 ASN485 SER486 ARG487 ASP488 GLN489 ARG492 LYS505 THR506 GLU507 GLY510 LYS511 ARG512 SER515 ALA516 ILE532 TYR535 ARG536 THR539 LYS540 LEU541 LYS542 SER543 THR544 TYR545 ILE546 ASP547 PRO548 ASP551 LEU552 ARG563 ASN565 ALA568 THR569 ALA570 SER577 ASP578 ASN580 ASN583 PRO585 ARG728 VAL730 PRO731 ASP732 ALA735 SER739 VAL740 ALA743 MET747 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)