Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 6006 |
| PDBID: | 8DLY |
| Chains: | H_C |
| Organism: | Severe acute respiratory syndrome coronavirus 2, Homo sapiens |
| Method: | EM |
| Resolution (Å): | 3.00 |
| Reference: | 10.1038/s41467-022-32262-8 |
| Antibody | |
| Antibody: | ab6 VHH |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | SARS-CoV-2 Epsilon (B.1.429) spike glycoprotein |
| Antigen mutation: | No |
| Durg Target: | P0DTC2 |
Sequence information
Antibody
Chain: H
Mutation: NULL
| >8DLY_H|Chain B[auth H]|VH ab6|Homo sapiens (9606) EVQLVESGGGVVQPGRSLRLSCAASGFTFSSYAMHWVRQAPGKGLEWIGNIYHDGSTFYNPSLKSLVTISRDDSTNTLYLQMNSLRAEDTAIYYCARVWLYGSGYMDVWGKGTLVTVSS |
Antigen
Chain: C
Mutation: NULL
| >8DLY_C|Chain A[auth C]|Spike glycoprotein|Severe acute respiratory syndrome coronavirus 2 (2697049) MFVFLVLLPLVSIQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSCMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYRYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQGVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPGSASSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSPIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGPALQIPFPMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTPSALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDPPEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQGSGYIPEAPRDGQAYVRKDGEWVLLSTFLGRSLEVLFQGPGHHHHHHHHSAWSHPQFEKGGGSGGGGSGGSAWSHPQFEK |
Interaction
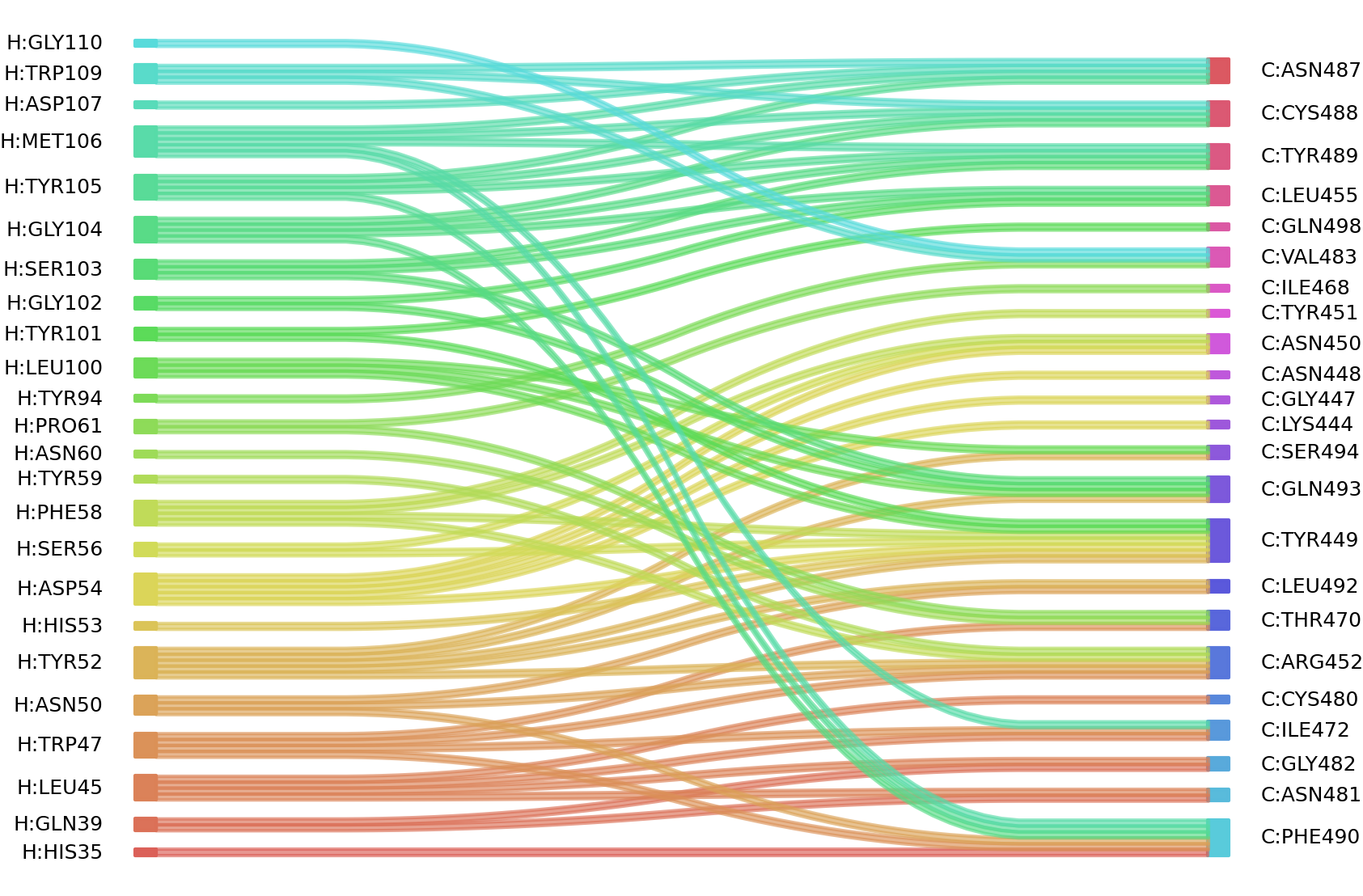
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition H: HIS35 VAL37 GLN39 GLY44 LEU45 TRP47 ASN50 TYR52 HIS53 ASP54 SER56 PHE58 TYR59 ASN60 PRO61 TYR94 TRP99 LEU100 TYR101 GLY102 SER103 GLY104 TYR105 MET106 ASP107 TRP109 GLY110 LYS111 C: SER349 TYR351 LYS444 GLY446 GLY447 ASN448 TYR449 ASN450 ARG452 LEU455 ILE468 THR470 GLU471 ILE472 CYS480 ASN481 GLY482 VAL483 PHE486 ASN487 CYS488 TYR489 PHE490 LEU492 GLN493 SER494 GLN498 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)