Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 5249 |
| PDBID: | 7UZB |
| Chains: | HL_A |
| Organism: | Severe acute respiratory syndrome coronavirus 2, Mus musculus |
| Method: | EM |
| Resolution (Å): | 4.10 |
| Reference: | 10.1016/j.immuni.2022.10.019 |
| Antibody | |
| Antibody: | HSW-2 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | SARS-CoV-2 spike glycoprotein |
| Antigen mutation: | Yes |
| Durg Target: | P0DTC2 |
Sequence information
Antibody
Heavy Chain: H
Mutation: NULL
| >7UZB_H|Chain B[auth H]|HSW-2 Fab heavy chain|Mus musculus (10090) QVQLQQSGPELVKPGASVKISCKASGYVFSTSWMSWVKQRPGEGPEWIGRIYPRDGHSSSTGKFKDKATLTADKSSNTAYIHLSSLTSEDSAVYFCARDYGYYYFDYWGQGTTLTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKRVEPKSCDKTHHHHHH |
Light Chain: L
Mutation: NULL
| >7UZB_L|Chain C[auth L]|HSW-2 Fab light chain|Mus musculus (10090) DIQMTQSPASLSASVGEAVTITCRLSENVYSFLAWYQQKQGKSPQLLVYRAKTLAEGVPSRFSGSGSGTQFSLKINSLQPEDFGTYYCQHHYGTPPTFGGGTKLEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC |
Antigen
Chain: A
Mutation: K814P/G889P/I896P/S939P/R983P/L984P
| >7UZB_A|Chain A|Spike glycoprotein|Severe acute respiratory syndrome coronavirus 2 (2697049) MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPASVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSPIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGPALQIPFPMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTPSALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDPPEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPSGRLVPRGSPGSGYIPEAPRDGQAYVRKDGEWVLLSTFLGHHHHHH |
Interaction
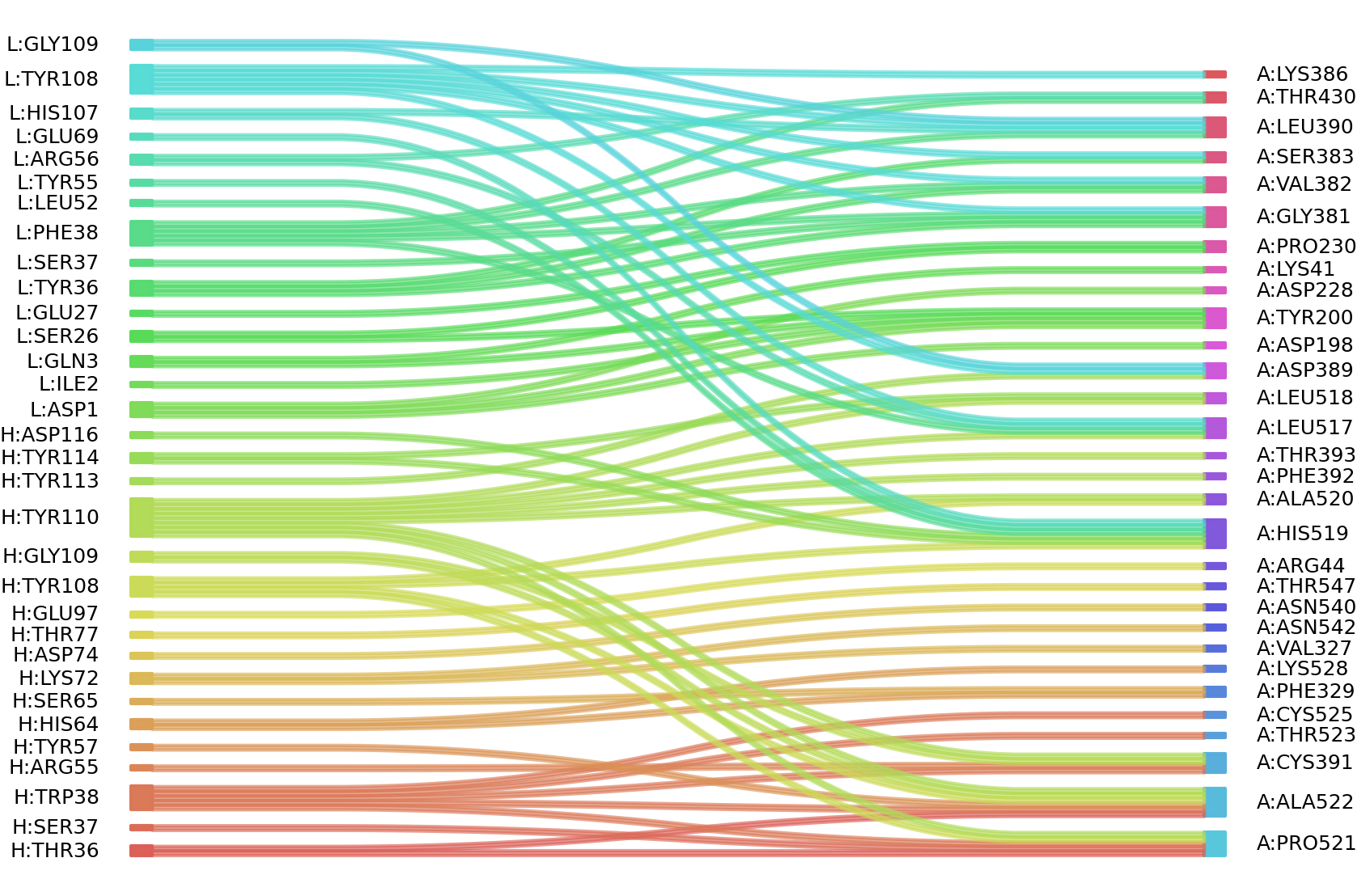
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition H: THR36 SER37 TRP38 ARG45 GLU48 ARG55 TYR57 ARG59 ASP62 HIS64 SER65 SER66 LYS72 ASP74 THR77 SER96 GLU97 TYR108 GLY109 TYR110 TYR113 TYR114 ASP116 L: ASP1 GLN3 SER26 GLU27 TYR36 SER37 PHE38 LEU52 TYR55 ARG56 GLU69 HIS107 TYR108 GLY109 THR114 A: LYS41 VAL42 ARG44 VAL47 HIS49 PHE168 ASP198 TYR200 LYS202 ASP228 PRO230 SER325 VAL327 PHE329 ASN360 GLY381 VAL382 SER383 LYS386 ASP389 LEU390 CYS391 PHE392 THR393 ASP428 THR430 LEU517 LEU518 HIS519 ALA520 PRO521 ALA522 THR523 CYS525 LYS528 ASN540 ASN542 THR547 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)