Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 5008 |
| PDBID: | 7TDN |
| Chains: | HL_B |
| Organism: | Homo sapiens, Clostridium perfringens |
| Method: | EM |
| Resolution (Å): | 5.00 |
| Reference: | 10.1016/j.jbc.2022.102357 |
| Antibody | |
| Antibody: | COP-3 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | Heat-labile enterotoxin B chain (cCpE) |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Heavy Chain: H
Mutation: NULL
| >7TDN_H|Chain C[auth H]|COP-3 Fab Heavy chain|Homo sapiens (9606) EISEVQLVESGGGLVQPGGSLRLSCAASGFNFYSSSIHWVRQAPGKGLEWVAYISSYSGYTYYADSVKGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARGYGYFDYNFSVGYALDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHT |
Light Chain: L
Mutation: NULL
| >7TDN_L|Chain D[auth L]|COP-3 Fab Light chain|Homo sapiens (9606) SDIQMTQSPSSLSASVGDRVTITCRASQSVSSAVAWYQQKPGKAPKLLIYSASSLYSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQSHPWYYPITFGQGTKVEIKRTVAAPSVFIFPPSDSQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC |
Antigen
Chain: B
Mutation: NULL
| >7TDN_B|Chain B|Heat-labile enterotoxin B chain|Clostridium perfringens (1502) MSTDIEKEILDLAAATERLNLTDALNSNPAGNLYDWRSSNSYPWTQKLNLHLTITATGQKYRILASKIVDFNIYSNNFNNLVKLEQSLGDGVKDHYVDISLDAGQYVLVMKANSSYSGNYPYSILFQKFGLVPR |
Interaction
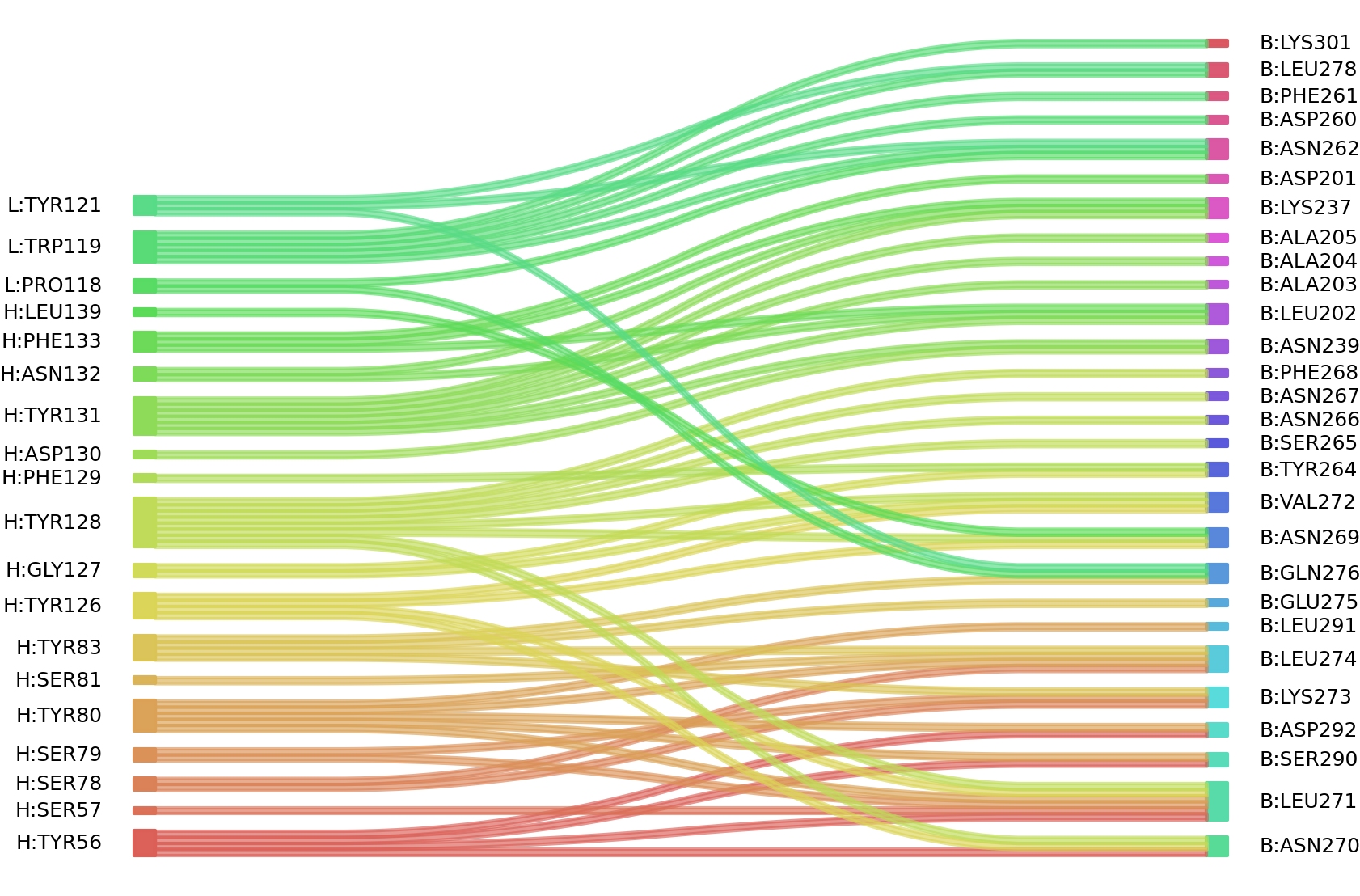
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition H: TYR56 SER57 SER59 SER78 SER79 TYR80 SER81 TYR83 GLY125 TYR126 GLY127 TYR128 PHE129 ASP130 TYR131 ASN132 PHE133 SER134 LEU139 L: PRO118 TRP119 TYR121 B: LEU200 ASP201 LEU202 ALA203 ALA205 LYS237 ASN239 ASP260 PHE261 ASN262 TYR264 SER265 ASN266 ASN269 ASN270 LEU271 VAL272 LYS273 LEU274 GLU275 GLN276 LEU278 ILE289 SER290 LEU291 ASP292 VAL297 VAL299 LYS301 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)