Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 4914 |
| PDBID: | 7SO7 |
| Chains: | XY_B |
| Organism: | Clostridioides difficile, Homo sapiens |
| Method: | XRD |
| Resolution (Å): | 3.59 |
| Reference: | 10.1016/j.crstbi.2022.03.003 |
| Antibody | |
| Antibody: | B1 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | TcdB GTD |
| Antigen mutation: | No |
| Durg Target: | P18177 |
Sequence information
Antibody
Heavy Chain: X
Mutation: NULL
| >7SO7_X|Chain C[auth H], E[auth X]|Fab B1 HC|Homo sapiens (9606) EVQLVESGGGLVQPGGSLRLSCAASGFTFRSYWMHWVRQVPGKGLVWVSCINKEGSSTTYADSVKGRFTISRDNAKNTLYLEMNSLRADDTAVYYCLRGYDVDYWGQGTLVTVSSASTKGPSVFPLAPGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCD |
Light Chain: Y
Mutation: NULL
| >7SO7_Y|Chain D[auth L], F[auth Y]|FAB B1 LC|Homo sapiens (9606) QAVLTQPSSLSASPGASVSLTCTLRSAINVGTYRIYWYQQKPGSPPRYLLRYKSGLDKHQGSGVPSRFSGSKDDSANAGILFISGLQSEDEADYYCLIWHSSAVVFGGGTKLTVLGQPKAAPSVTLFPPSSEELQANKATLVCLISDFYPGAVTVAWKADGSPVKAGVETTKPSKQSNNKYAASSYLSLTPEQWKSHRSYSCQVTHEGSTVEKTVAPT |
Antigen
Chain: B
Mutation: NULL
| >7SO7_B|Chain A, B|Toxin B|Clostridioides difficile (1496) MSLVNRKQLEKMANVRFRTQEDEYVAILDALEEYHNMSENTVVEKYLKLKDINSLTDIYIDTYKKSGRNKALKKFKEYLVTEVLELKNNNLTPVEKNLHFVWIGGQINDTAINYINQWKDVNSDYNVNVFYDSNAFLINTLKKTVVESAINDTLESFRENLNDPRFDYNKFFRKRMEIIYDKQKNFINYYKAQREENPELIIDDIVKTYLSNEYSKEIDELNTYIEESLNKITQNSGNDVRNFEEFKNGESFNLYEQELVERWNLAAASDILRISALKEIGGMYLDVDMLPGIQPDLFESIEKPSSVTVDFWEMTKLEAIMKYKEYIPEYTSEHFDMLDEEVQSSFESVLASKSDKSEIFSSLGDMEASPLEVKIAFNSKGIINQGLISVKDSYCSNLIVKQIENRYKILNNSLNPAISEDNDFNTTTNTFIDSIMAEANADNGRFMMELGKYLRVGFFPDVKTTINLSGPEAYAAAYQDLLMFKEGSMNIHLIEADLRNFEISKTNISQSTEQEMASLWSFDDARAKAQFEEYKRNY |
Interaction
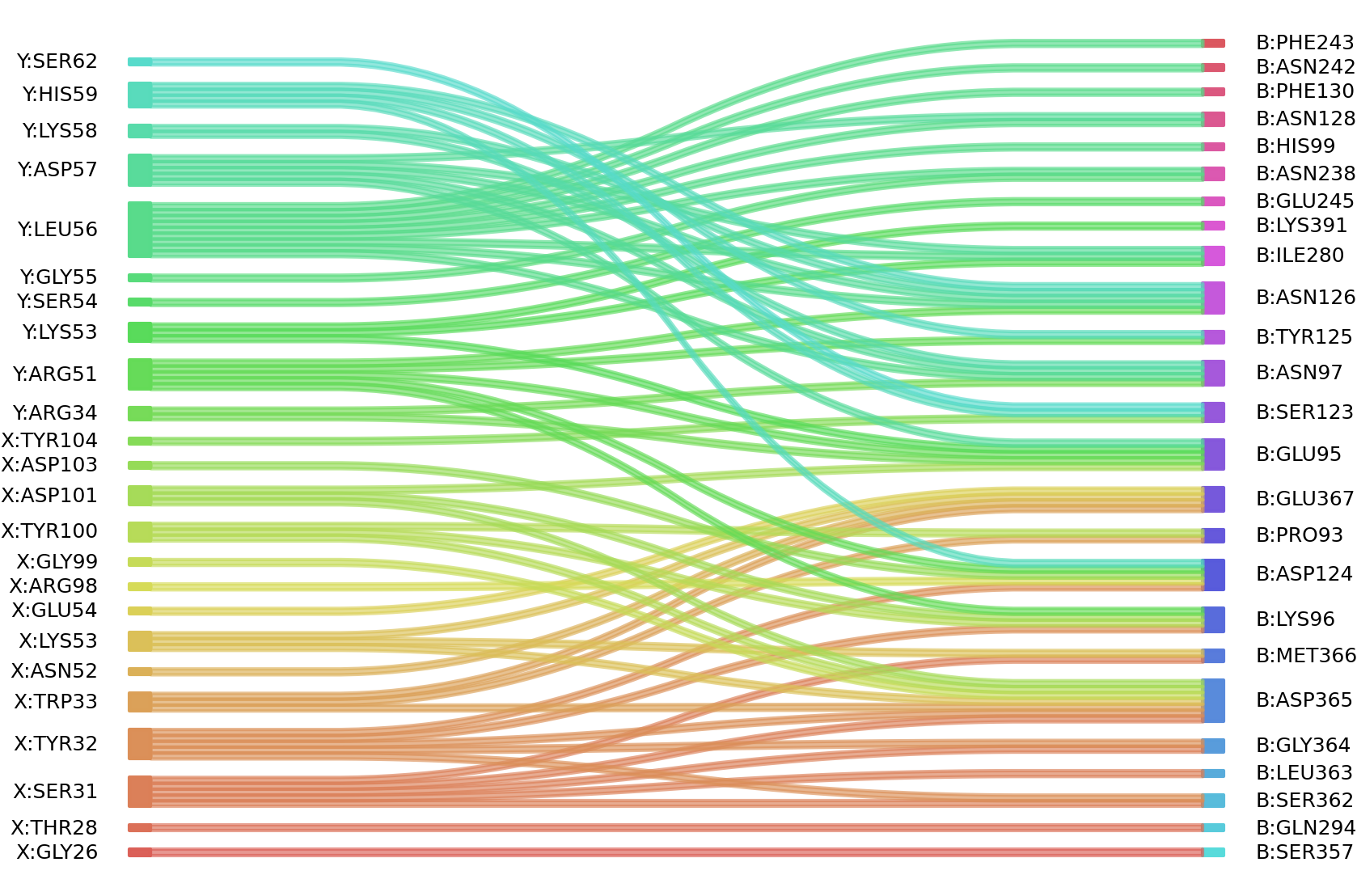
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition X: GLY26 THR28 SER31 TYR32 TRP33 ASN52 LYS53 GLU54 ARG98 TYR100 ASP101 ASP103 TYR104 Y: ARG34 ARG51 LYS53 SER54 GLY55 LEU56 ASP57 LYS58 HIS59 SER62 B: PRO93 VAL94 GLU95 LYS96 ASN97 SER123 ASP124 TYR125 ASN126 ASN128 PHE130 ASN238 ASP239 ASN242 PHE243 GLU245 ILE280 GLN294 SER357 SER362 LEU363 GLY364 ASP365 MET366 GLU367 LYS391 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)