Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 4068 |
| PDBID: | 7LAB |
| Chains: | TS_A |
| Organism: | Homo sapiens, Severe acute respiratory syndrome coronavirus 2 |
| Method: | EM |
| Resolution (Å): | 2.97 |
| Reference: | 10.1016/j.cell.2021.06.021 |
| Antibody | |
| Antibody: | DH1052 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | SARS-CoV-2 Spike glycoprotein |
| Antigen mutation: | No |
| Durg Target: | P0DTC2 |
Sequence information
Antibody
Heavy Chain: T
Mutation: NULL
| >7LAB_T|Chain B[auth H], D[auth T], H[auth Y]|DH1052 heavy chain|Homo sapiens (9606) EVQLVESGAEVKKPGATVKISCKVSGYTFTDYYMHWVQQAPGKGLEWMGLVDPEDGETIYAEKFQGRVTITADTSTDTAYMELSSLRSEDTAVYYCATSSGPSRLCGGGSCYHSFDYWGQGTLVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVE |
Light Chain: S
Mutation: NULL
| >7LAB_S|Chain A[auth L], C[auth S], I[auth X]|DH1052 light chain|Homo sapiens (9606) LTQSPGTLSLSPGERATLSCRASQSVSSSYLAWYQQKPGQAPRLLIYGASSRATGIPDRFSGSGSGTDFTLTISRLEPEDFAVYYCQQYGSSPTWTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQG |
Antigen
Chain: A
Mutation: NULL
| >7LAB_A|Chain E[auth A], F[auth B], G[auth C]|Spike glycoprotein|Severe acute respiratory syndrome coronavirus 2 (2697049) AYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDPPEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDS |
Interaction
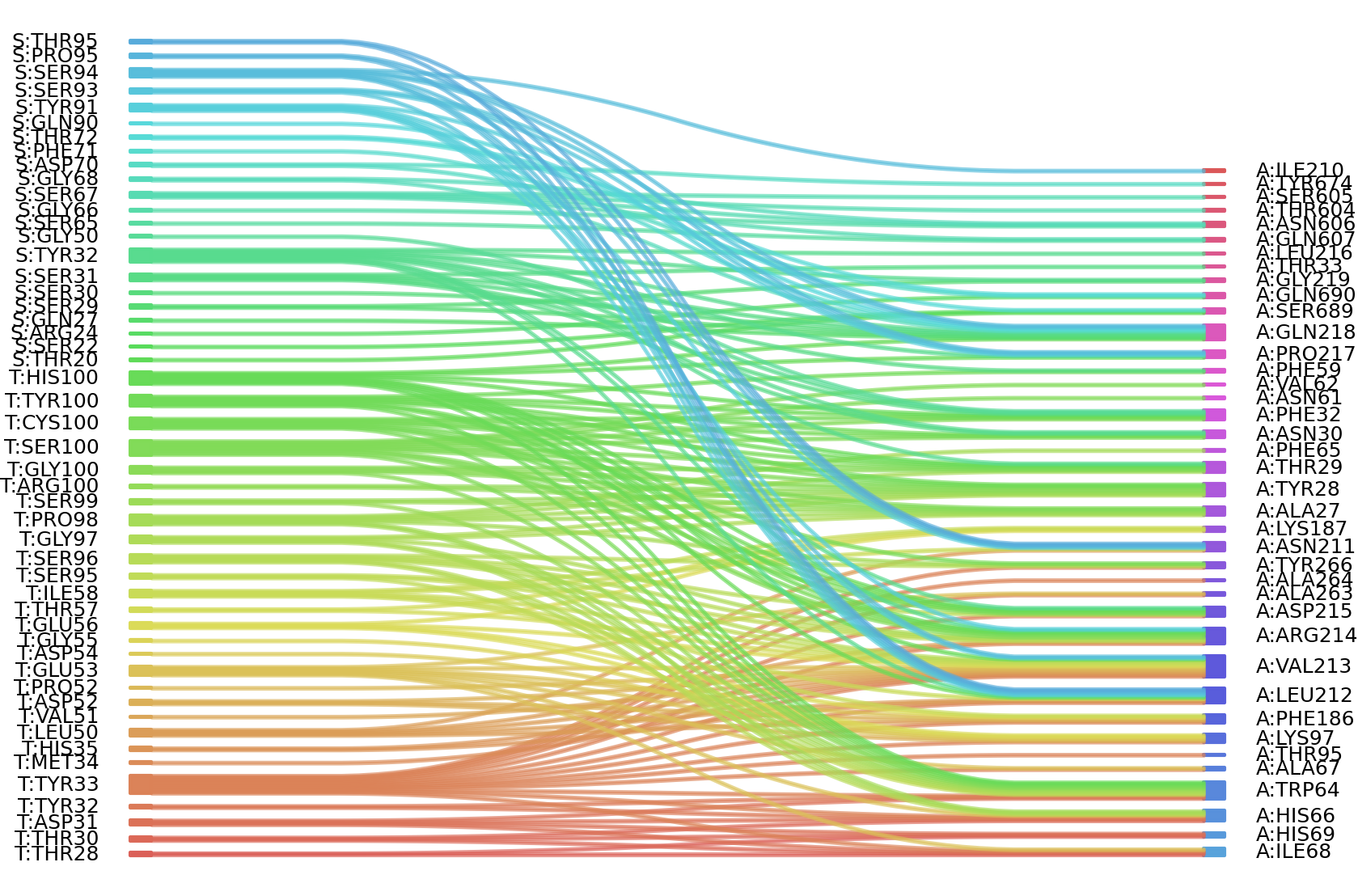
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition S: THR20 SER29 SER30 SER31 TYR32 SER67 THR72 TYR91 SER93 SER94 T: THR28 THR30 ASP31 TYR33 LEU50 GLU53 ASP54 GLU56 ILE58 SER95 SER96 PRO98 SER99 A: ALA27 TYR28 THR29 ASN30 PHE32 PHE59 ASN61 TRP64 HIS66 ILE68 HIS69 LYS97 PHE186 LYS187 ILE210 ASN211 LEU212 VAL213 ARG214 ASP215 PRO217 GLN218 ALA263 SER605 ASN606 SER689 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)