Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 3715 |
| PDBID: | 7EPU |
| Chains: | A_B |
| Organism: | Homo sapiens |
| Method: | XRD |
| Resolution (Å): | 3.50 |
| Reference: | 10.1038/s41467-021-24320-4 |
| Antibody | |
| Antibody: | 7EPU scFv |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | chromodomain helicase/ATPase DNA-binding protein 1-like(CHD1L) |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Chain: A
Mutation: NULL
| >7EPU_A|Chain A|non-immunized human scFv|Homo sapiens (9606) ASQVQLQQSGPGLVRPSQTLSLTCAISGDSVSDNDAAWNWVRQSPSRGLEWLGRTYYRSKWSNEYAVFVQSRITINPDTSKNQFSLHLNSVTPEDTAVYYCARATRPTGNGLDIWGQGTMVTVSSGILGSGGGGSGGGGSGGGGSSYVLTQPPSVSVAPGQTARITCGGNNIGSKSVHWYQQKPGQAPVLVVYDDSDRPSGIPERFSGSNSGNTAALTISRVEAGDEADYYCQVWDSSSDHVVFGGGTKLTVLSGILHHHHHH |
Antigen
Chain: B
Mutation: NULL
| >7EPU_B|Chain B|Chromodomain-helicase-DNA-binding protein 1-like|Homo sapiens (9606) MERAGATSRGGQAPGFLLRLHTEGRAEAARVQEQDLRQWGLTGIHLRSYQLEGVNWLAQRFHCQNGCILGDEMGLGKTCQTIALFIYLAGRLNDEGPFLILCPLSVLSNWKEEMQRFAPGLSCVTYAGDKEERACLQQDLKQESRFHVLLTTYEICLKDASFLKSFPWSVLVVDEAHRLKNQSSLLHKTLSEFSVVFSLLLTGTPIQNSLQELYSLLSFVEPDLFSKEEVGDFIQRYQDIEKESESASELHKLLQPFLLRRVKAEVATELPKKTEVVIYHGMSALQKKYYKAILMKDLDAFENETAKKVKLQNILSQLRKCVDHPYLFDGVEPEPFEVGDHLTEASGKLHLLDKLLAFLYSGGHRVLLFSQMTQMLDILQDYMDYRGYSYERVDGSVRGEERHLAIKNFGQQPIFVFLLSTRAGGVGMNLTAADTVIFVDSDFNPQNDLQAAARAHRIGQNKSVKVIRLIGRDTVEEIVYRKAASKLQLTNMIIEGGHFTLGAQKPAADADLQLSEILKFGLDKLLASEGSTMDEIDLESILGETKDGQWVSDALPAAEGGSRDQEEGKNHMYLFEGKDYSKEPSKEDRKSFEQLVNLQKTLLEKASQEGRSLRNKGSVLIPGLVEGSTKRKRVLSPEELEDRQKKRQEAAAKRRRLIEEKKRQKEEAEHKKKMAWWESNNYQSFCLPSEESEPEDLENGEESSAELDYQDPDATSLKYVSGDVTHPQAGAEDALIVHCVDDSGHWGRGGLFTALEKRSAEPRKIYELAGKMKDLSLGGVLLFPVDDKESRNKGQDLLALIVAQHRDRSNVLSGIKMAALEEGLKKIFLAAKKKKASVHLPRIGHATKGFNWYGTERLIRKHLAARGIPTYIYYFPRSKS |
Interaction
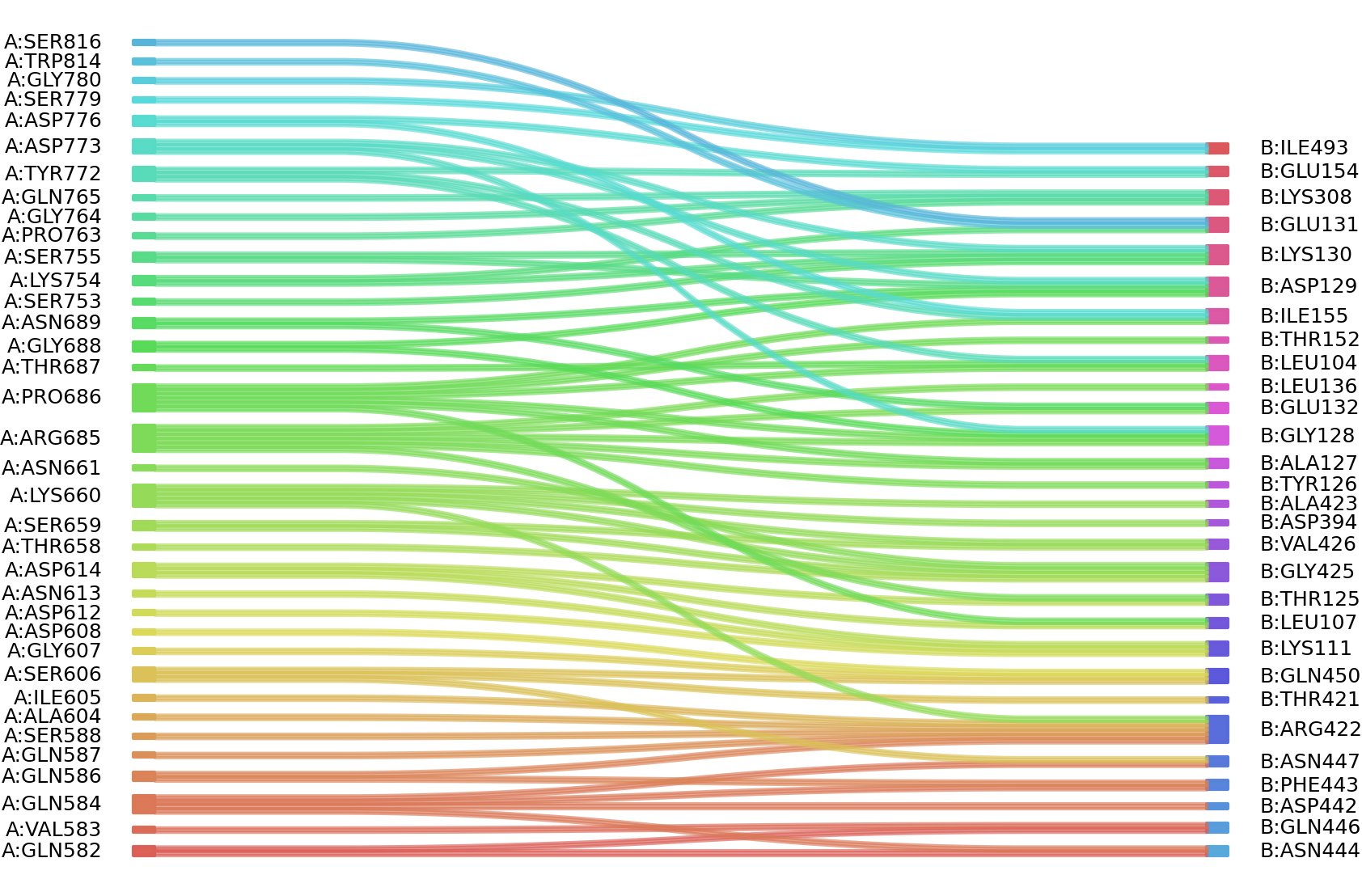
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition A: GLN582 VAL583 GLN584 GLN586 GLN587 SER588 ALA604 ILE605 SER606 GLY607 ASP608 ASP612 ASN613 ASP614 ARG637 THR658 SER659 LYS660 ASN661 THR684 ARG685 PRO686 THR687 GLY688 ASN689 ASP692 GLN696 GLY752 SER753 LYS754 SER755 PRO763 GLY764 GLN765 TYR772 ASP773 ASP776 PRO778 SER779 GLY780 TRP814 SER816 B: LEU104 SER105 LEU107 SER108 LYS111 THR125 ALA127 GLY128 ASP129 LYS130 GLU131 GLU132 LEU136 THR152 GLU154 ILE155 LYS158 LYS308 ASP394 THR421 ARG422 ALA423 GLY425 VAL426 ASP442 PHE443 ASN444 GLN446 ASN447 GLN450 ILE493 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)