Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 3563 |
| PDBID: | 7D30 |
| Chains: | A_B |
| Organism: | synthetic construct, Severe acute respiratory syndrome coronavirus 2 |
| Method: | XRD |
| Resolution (Å): | 2.10 |
| Reference: | 10.1371/journal.ppat.1009328 |
| Antibody | |
| Antibody: | MR17-SR31 synthetic-scFv |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | SARS-CoV-2 Spike glycoprotein RBD |
| Antigen mutation: | No |
| Durg Target: | P0DTC2 |
Sequence information
Antibody
Chain: A
Mutation: NULL
| >7D30_A|Chain A|sybody fusion of MR17-SR31 with a GS linker|synthetic construct (32630) GSSSQVQLVESGGGLVQAGGSLRLSCAASGFPVEVWRMEWYRQAPGKEREGVAAIESYGHGTRYADSVKGRFTISRDNAKNTVYLQMNSLKPEDTAVYYCNVKDDGQLAYHYDYWGQGTQVTVSGGGGSGGGGSGGGGSGGGGSGGGGSGGGGSGSSSQVQLVESGGGLVQAGGSLRLSCAASGFPVWQGEMAWYRQAPGKEREWVAAISSMGYKTYYADSVKGRFTISRDNAKNTVYLQMNSLKPEDTAVYYCAVMVGFWYAGQGTQVTVSAGRAGEQKLISEEDLNSAVDHHHHHH |
Antigen
Chain: B
Mutation: NULL
| >7D30_B|Chain B|Spike protein S1|Severe acute respiratory syndrome coronavirus 2 (2697049) AGSPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTGTLEVLFQ |
Interaction
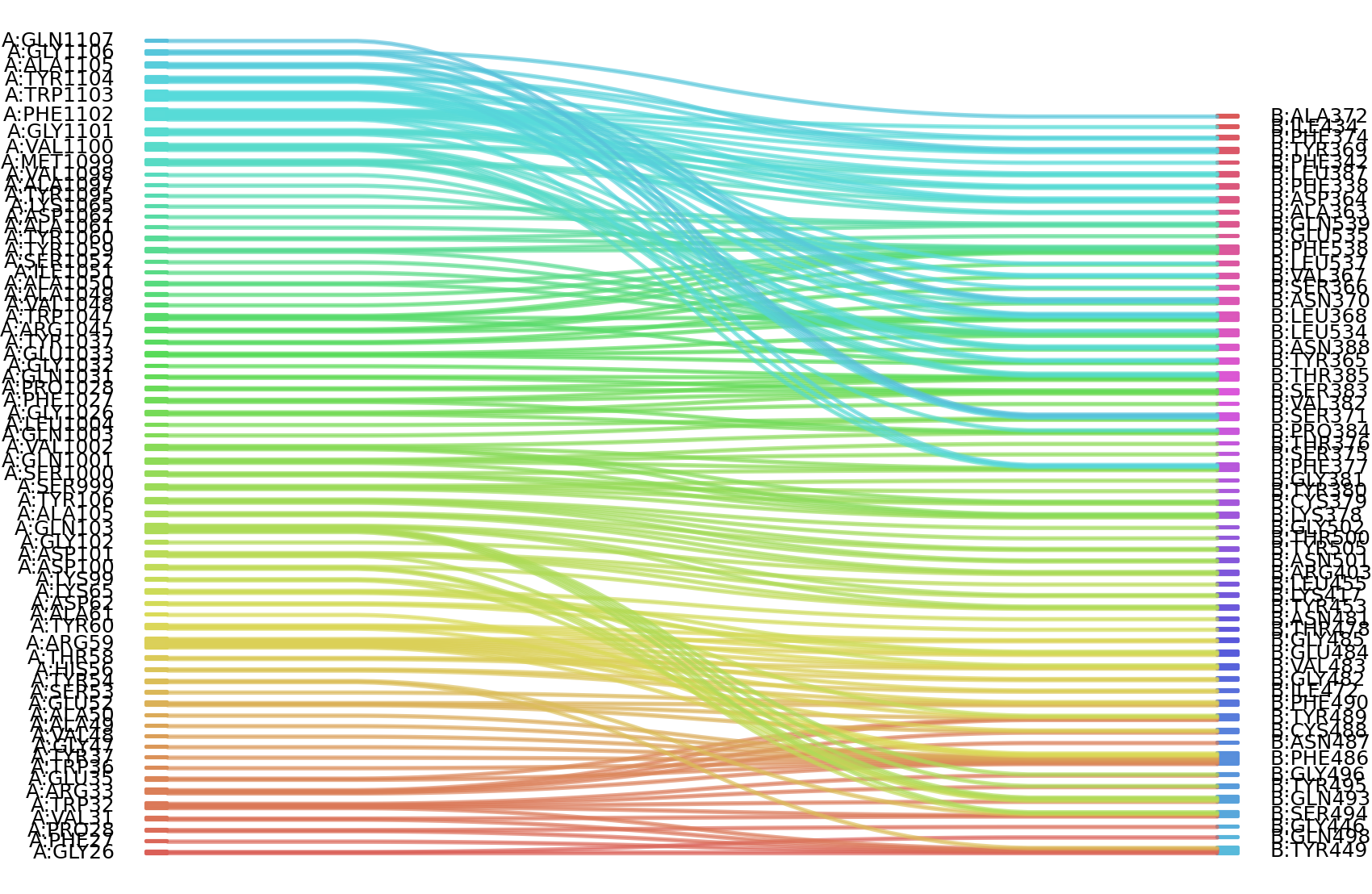
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition A: VAL2 GLY26 PHE27 PRO28 VAL31 TRP32 ARG33 GLU35 TYR37 GLY47 VAL48 ALA49 ALA50 GLU52 SER53 TYR54 HIS56 THR58 ARG59 TYR60 ALA61 ASP62 LYS65 LYS99 ASP101 GLY102 GLN103 LEU104 ALA105 TYR106 SER999 SER1000 GLN1001 VAL1002 GLN1003 LEU1004 GLY1026 PHE1027 PRO1028 GLN1031 GLY1032 GLU1033 TYR1037 ARG1045 TRP1047 VAL1048 ALA1049 ALA1050 ILE1051 SER1052 SER1053 TYR1059 TYR1060 ALA1061 ASP1062 LYS1065 TYR1095 VAL1098 MET1099 VAL1100 GLY1101 PHE1102 TRP1103 TYR1104 ALA1105 GLY1106 GLN1107 B: PHE338 PHE342 ALA363 ASP364 TYR365 SER366 VAL367 LEU368 TYR369 ASN370 SER371 PHE374 SER375 THR376 PHE377 LYS378 CYS379 TYR380 GLY381 VAL382 SER383 PRO384 THR385 LEU387 ASN388 ARG403 GLU406 LYS417 ILE434 GLY446 TYR449 LEU452 TYR453 LEU455 PHE456 ILE472 THR478 ASN481 GLY482 VAL483 GLU484 GLY485 PHE486 CYS488 TYR489 PHE490 LEU492 GLN493 SER494 TYR495 GLY496 PHE497 GLN498 THR500 ASN501 GLY502 TYR505 LEU513 THR533 LEU534 GLU535 LEU537 PHE538 GLN539 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)