Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 2581 |
| PDBID: | 6OAO |
| Chains: | H_G |
| Organism: | Plasmodium vivax, Homo sapiens |
| Method: | XRD |
| Resolution (Å): | 3.50 |
| Reference: | 10.1038/s41564-019-0461-2 |
| Antibody | |
| Antibody: | 092096 scFv |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | Duffy binding surface protein region II |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Chain: H
Mutation: NULL
| >6OAO_H|Chain B, D, F, H, J, L|Antibody 092096 single chain variable fragment|Homo sapiens (9606) MVHSQVQLVQSGAEVKKPGASVKVSCKVSGYTLTELSMHWVRQAPGKGLEWMGGFDREDGESIYAQKFQGRVTLTEDTSTDSVYMELSNLRSDDTAVYFCATDFGGSYFYAFDIWGQGTMVTVSSGGGGSGGGGSGGGGSQSVLTQSPSASGTPGQRVTISCSGSSSNIGRNPVNWFQHLPGTAPQLLIYSNDQRPSGVPDRFSGSKSGTSASLAISGLQSEDEADYYCEAWDDSLNGVVFGGGTKLTVLG |
Antigen
Chain: G
Mutation: NULL
| >6OAO_G|Chain A, C, E, G, I, K|Duffy binding surface protein region II|Plasmodium vivax (5855) NTVMKNCNYKRKRRERDWDCNTKKDVCIPDRRYQLCMKELTNLVNNTDTNFHRDITFRKLYLKRKLIYDAAVEGDLLLKLNNYRYNKDFCKDIRWSLGDFGDIIMGTDMEGIGYSKVVENNLRSIFGTDEKAQQRRKQWWNESKAQIWTAMMYSVKKRLKGNFIWICKLNVAVNIEPQIYRWIREWGRDYVSELPTEVQKLKEKCDGKINYTDKKVCKVPPCQNACKSYDQWITRKKNQWDVLSNKFISVKNAEKVQTAGIVTPYDILKQELDEFNEVAFENEINKRDGAYIELCVCSVEEAKKNTQEVVTNVDN |
Interaction
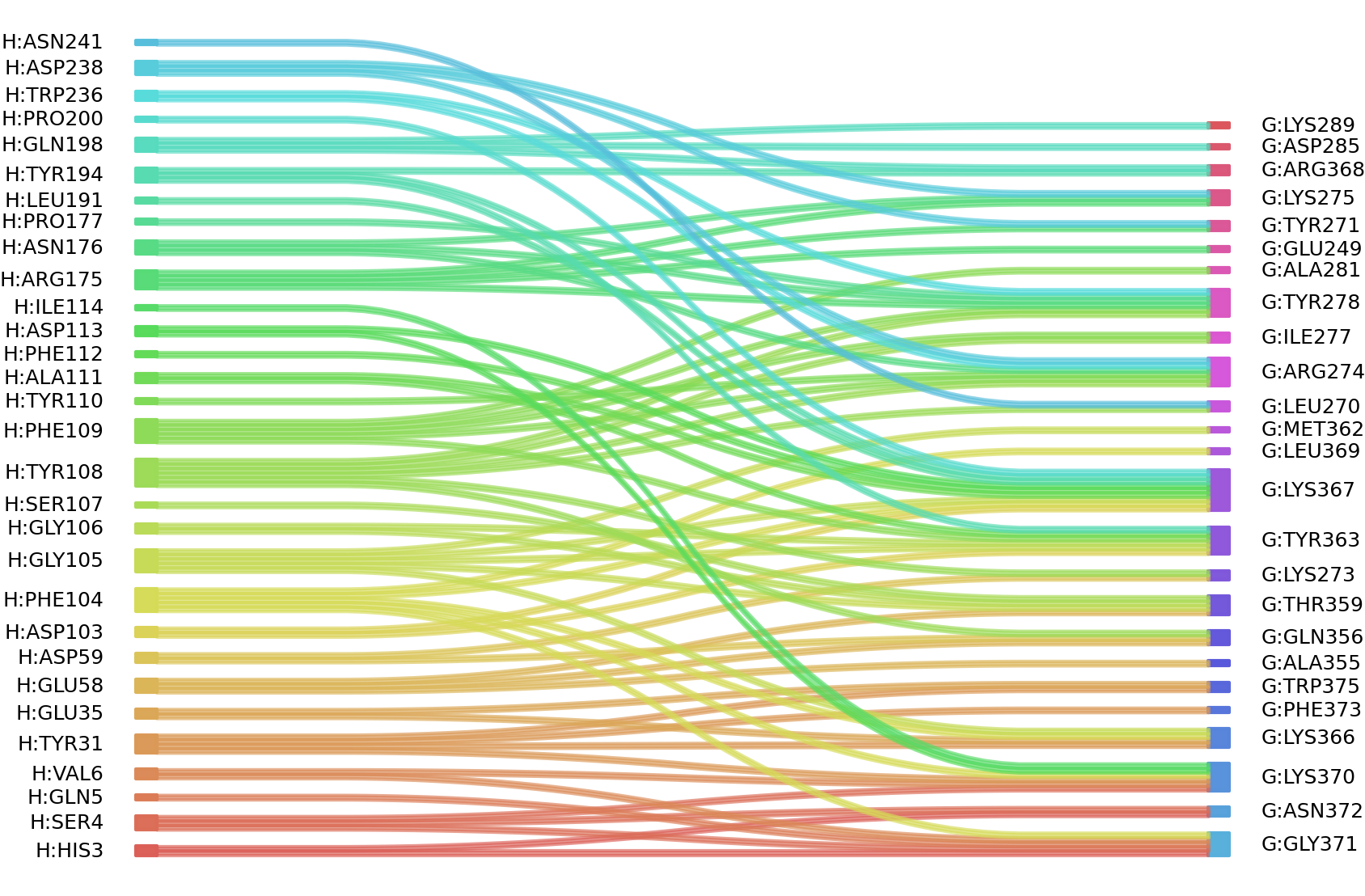
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition H: HIS3 SER4 GLN5 VAL6 GLY30 TYR31 GLU35 ASP56 GLU58 ASP59 ASP103 PHE104 GLY105 GLY106 SER107 TYR108 PHE109 TYR110 ALA111 ASP113 ILE114 ARG175 ASN176 PRO177 LEU191 TYR194 SER195 GLN198 ARG199 PRO200 SER201 TRP236 ASP238 ASN241 G: GLU249 LEU270 TYR271 LYS273 ARG274 LYS275 ILE277 TYR278 ALA281 VAL282 ASP285 LYS289 GLN356 THR359 ALA360 TYR363 SER364 LYS366 LYS367 ARG368 LEU369 LYS370 GLY371 ASN372 PHE373 TRP375 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)