Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 2504 |
| PDBID: | 6MWV |
| Chains: | KL_J |
| Organism: | Eastern equine encephalitis virus, Mus musculus |
| Method: | EM |
| Resolution (Å): | 7.30 |
| Reference: | 10.1016/j.celrep.2018.11.067 |
| Antibody | |
| Antibody: | EEEV-58 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | chimeric Eastern Equine Encephalitis Virus E2 |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Heavy Chain: K
Mutation: NULL
| >6MWV_K|Chain C, G, K, O|EEEV-58 antibody heavy chain|Mus musculus (10090) QIQLVQSGREVKNPGETVKISCKASGYTFTEYPMLWVKQAPGKGFRWMGLIYTNTGEPTYAEEFKGRFVFSLEISASTAYLQINNLTNEDTATYFCVRDYFISLDYWGQGTTLTVSSAKTTAPSVYPLAPVCGGTTGSSVTLGCLVKGYFPEPVTLTWNSGSLSSGVHTFPALLQSGLYTLSSSVTVTSNTWPSQTITCNVAHPASSTKVDKKIESRR |
Light Chain: L
Mutation: NULL
| >6MWV_L|Chain D, H, L, P|EEEV-58 antibody light chain|Mus musculus (10090) QAVVTQESALTTSPGETVTLTCRSNIGAVTSSNCANWVQEKPDHFFTGLIGDTNNRRSGVPARFSGSLIGDKAALTITGAQTEDEAIYFCALWYNNLWVFGGGTKLTVLGQPKSSPSVTLFPPSSEELETNKATLVCTITDFYPGVVTVDWKVDGTPVTQGMETTQPSKQSNNKYMASSYLTLTARAWERHSSYSCQVTHEGHTVEKSLSRADC |
Antigen
Chain: J
Mutation: NULL
| >6MWV_J|Chain B, F, J, N|E2|Eastern equine encephalitis virus (11021) DLDTHFTQYKLARPYIADCPNCGHSRCDSPIAIEEVRGDAHAGVIRIQTSAMFGLKTDGVDLAYMSFMNGKTQKSIKIDNLHVRTSAPCSLVSHHGYYILAQCPPGDTVTVGFHDGPNRHTCTVAHKVEFRPVGREKYRHPPEHGVELPCNRYTHKRADQGHYVEMHQPGLVADHSLLSIHSAKVKITVPSGAQVKYYCKCPDVREGITSSDHTTTCTDVKQCRAYLIDNKKWVYNSGRLPRGEGDTFKGKLHVPFVPVKAKCIATLAPEPLVEHKHRTLILHLHPDHPTLLTTRSLGSDANPTRQWIERPTTVNFTVTGEGLEYTWGNHPPKRVWAQESGEGNPHGWPHEVVVYYYNRYPLTTIIGLCTCVAIIMVSCVTSVWLLCRTRNLCITPYKLAPNAQVPILLALLCCIKPTRA |
Interaction
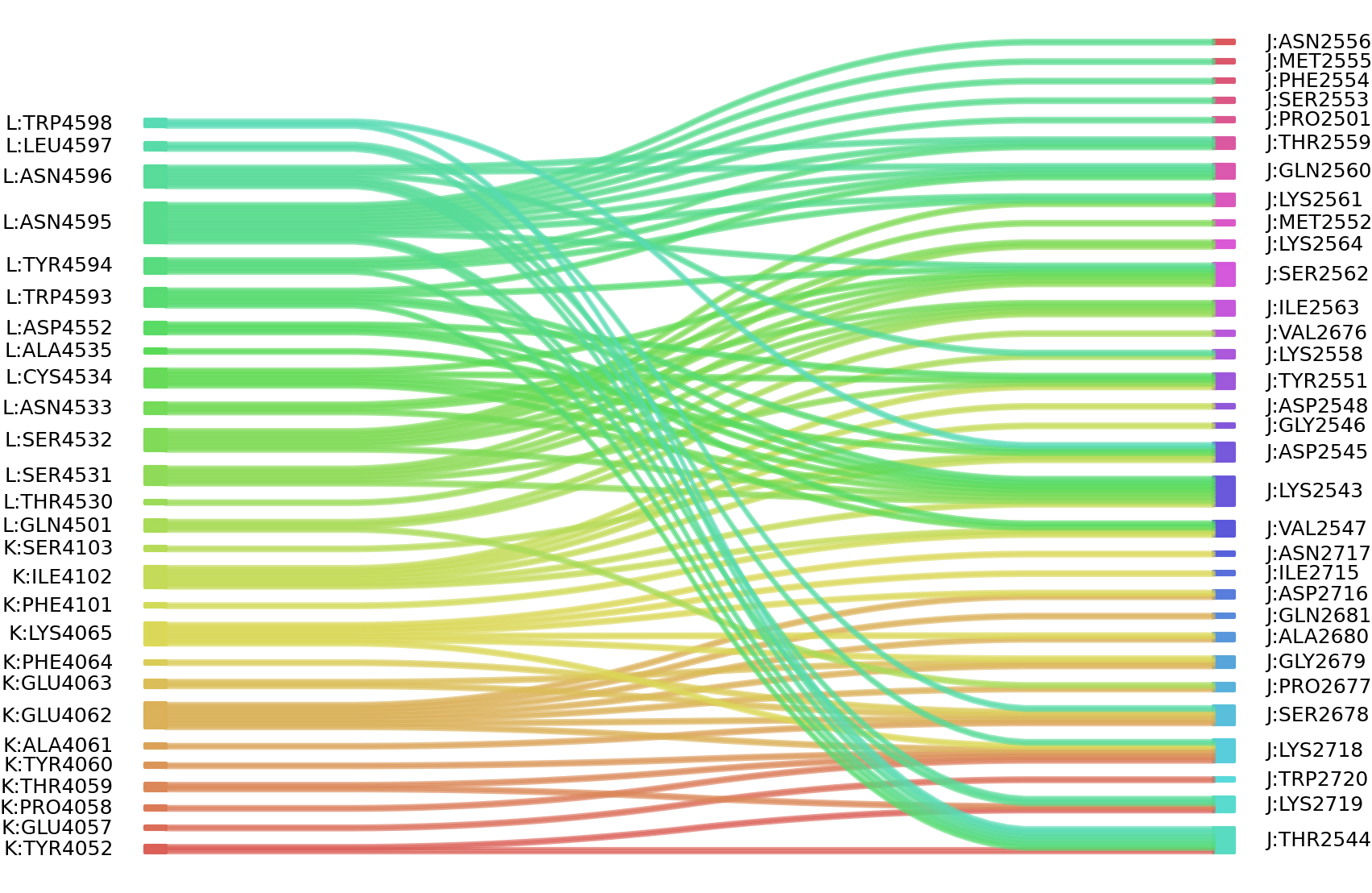
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition K: LEU4050 TYR4052 GLU4057 PRO4058 THR4059 TYR4060 ALA4061 GLU4062 GLU4063 LYS4065 ASP4099 PHE4101 ILE4102 SER4103 L: GLN4501 ILE4526 THR4530 SER4531 SER4532 ASN4533 CYS4534 ASP4552 TRP4593 TYR4594 ASN4595 ASN4596 LEU4597 TRP4598 J: LYS2543 THR2544 ASP2545 GLY2546 VAL2547 ASP2548 TYR2551 LYS2558 THR2559 GLN2560 LYS2561 SER2562 ILE2563 LYS2564 THR2675 VAL2676 PRO2677 SER2678 GLY2679 ALA2680 GLN2681 ILE2715 ASP2716 ASN2717 LYS2718 LYS2719 TRP2720 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)