Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 191 |
| PDBID: | 1YNT |
| Chains: | BA_F |
| Organism: | Mus musculus, Finegoldia magna ATCC 29328, Toxoplasma gondii |
| Method: | XRD |
| Resolution (Å): | 3.10 |
| Reference: | 10.1016/j.jmb.2005.09.028 |
| Antibody | |
| Antibody: | 4F11E12 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | SAG1 surface antigen (P30) |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Heavy Chain: B
Mutation: NULL
| >1YNT_B|Chain B, D|4F11E12 Fab variable heavy chain region|Mus musculus (10090) QVQLQQSGAELVRPGVSVKISCKGSGYTFTDYGMHWVKQSHAKSLEWIGIISTYSGDASYNQKFKGKATMTVDKSSSTAYMELARLTSEDSAIYYCARSSTWYYFDYWGQGTTLTVSSAKTTAPSVYPLAPVCGDTTGSSVTLGCLVKGYFPEPVTLTWNSGSLSSGVHTFPAVLQSDLYTLSSSVTVTSSTWPSQSITCNVAHPASSTKVDKKIEPR |
Light Chain: A
Mutation: NULL
| >1YNT_A|Chain A, C|4F11E12 Fab variable light chain region|Mus musculus (10090) DIQVTQTTSSLSASLGDRVTISCRASQDISNYLNWYQQKPDGTVKLLIYYTSRLHSGVPSRFSGSGSGTDYSLTISNLEQEDIATYFCQQGNTLPYTFGGGTKLEIKRADAAPTVSIFPPSSEQLTSGGASVVCFLNNFYPKDINVKWKIDGSERQNGVLNSWTDQDSKDSTYSMSSTLTLTKDEYERHNSYTCEATHKTSTSPIVKSFNRNE |
Antigen
Chain: F
Mutation: NULL
| >1YNT_F|Chain F, G|Major surface antigen p30|Toxoplasma gondii (5811) PPLVANQVVTCPDKKSTAAVILTPTENHFTLKCPKTALTEPPTLAYSPNRQICPAGTTSSCTSKAVTLSSLIPEAEDSWWTGDSASLDTAGIKLTVPIEKFPVTTQTFVVGCIKGDDAQSCMVTVTVQARASSVVNNVARCSYGADSTLGPVKLSAEGPTTMTLVCGKDGVKVPQDNNQYCSGTTLTGCNEKSFKDILPKLTENPWQGNASSDKGATLTIKKEAFPAESKSVIIGCTGGSPEKHHCTVKLEFAG |
Interaction
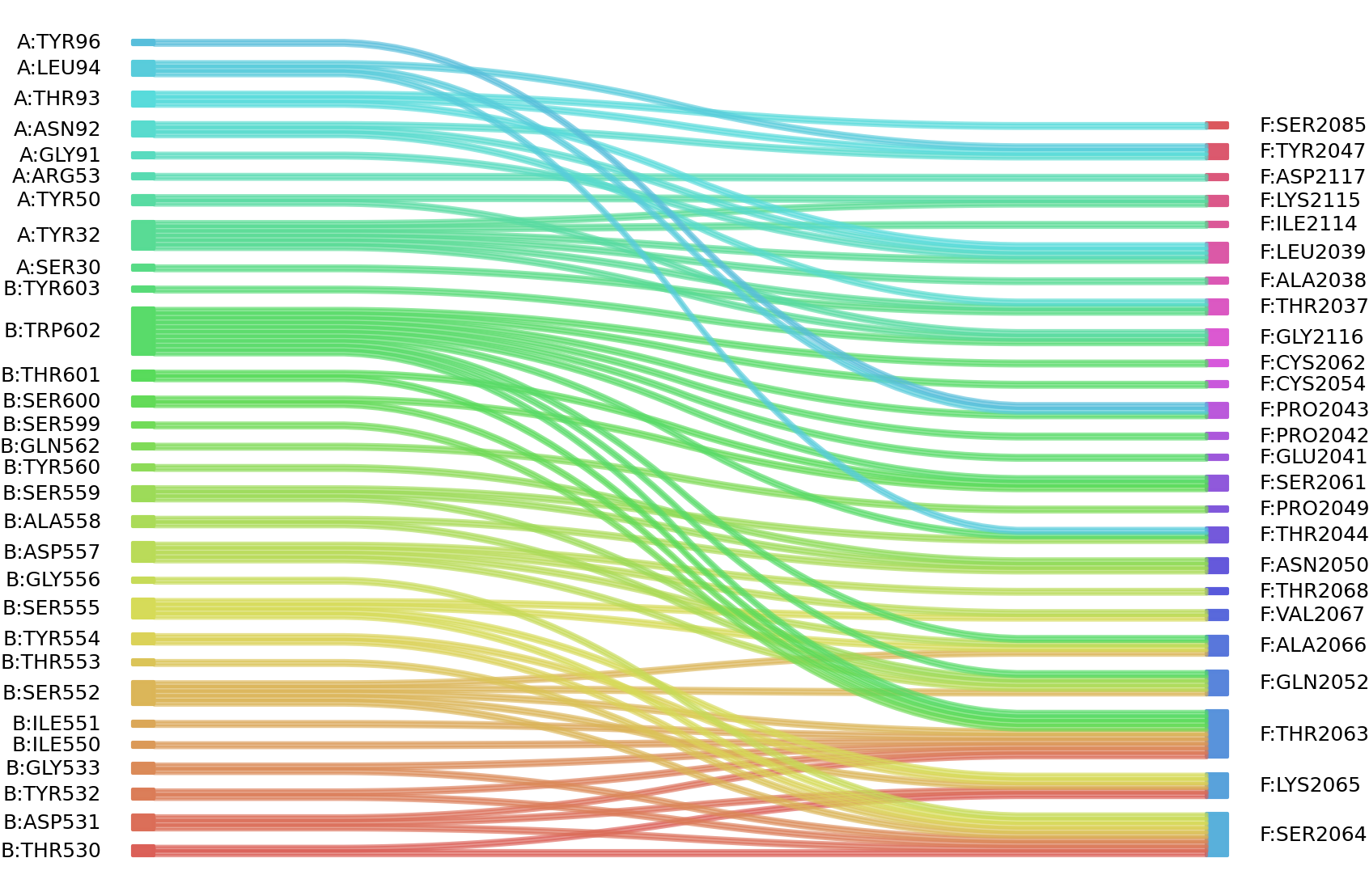
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition A: SER30 TYR32 TYR50 ARG53 GLY91 ASN92 THR93 LEU94 TYR96 B: THR530 ASP531 TYR532 GLY533 ILE550 SER552 THR553 TYR554 SER555 ASP557 ALA558 SER559 TYR560 GLN562 SER599 SER600 THR601 TRP602 TYR603 F: THR2037 ALA2038 LEU2039 GLU2041 PRO2042 PRO2043 THR2044 TYR2047 PRO2049 ASN2050 GLN2052 SER2061 CYS2062 THR2063 SER2064 LYS2065 ALA2066 VAL2067 THR2068 SER2085 ILE2114 LYS2115 GLY2116 ASP2117 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)