Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 145 |
| PDBID: | 1QFW |
| Chains: | IM_B |
| Organism: | Homo sapiens, Mus musculus |
| Method: | XRD |
| Resolution (Å): | 3.50 |
| Reference: | 10.1006/jmbi.1999.2845 |
| Antibody | |
| Antibody: | 3468 Fv |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | gonadotropin beta subunit |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Heavy Chain: I
Mutation: NULL
| >1QFW_I|Chain F[auth I]|ANTIBODY (ANTI BETA SUBUNIT) (HEAVY CHAIN)|Mus musculus (10090) QVQLQESGGHLVKPGGSLKLSCAASGFAFSSFDMSWIRQTPEKRLEWVASITNVGTYTYYPGSVKGRFSISRDNARNTLNLQMSSLRSEDTALYFCARQGTAAQPYWYFDVWGAGTTVTVSS |
Light Chain: M
Mutation: NULL
| >1QFW_M|Chain E[auth M]|ANTIBODY (ANTI BETA SUBUNIT) (LIGHT CHAIN)|Mus musculus (10090) DIELTQSPKSMSMSVGERVTLSCKASETVDSFVSWYQQKPEQSPKLLIFGASNRFSGVPDRFTGSGSATDFTLTISSVQAEDFADYHCGQTYNHPYTFGGGTKLEIKR |
Antigen
Chain: B
Mutation: NULL
| >1QFW_B|Chain B|GONADOTROPHIN BETA SUBUNIT|Homo sapiens (9606) SKEPLRPRCRPINATLAVEKEGCPVCITVNTTICAGYCPTMTRVLQGVLPALPQVVCNYRDVRFESIRLPGCPRGVNPVVSYAVALSCQCALCRRSTTDCGGPKDHPLTCDDPRFQDSSSSKAPPPSLPSPSRLPGPSDTPILPQ |
Interaction
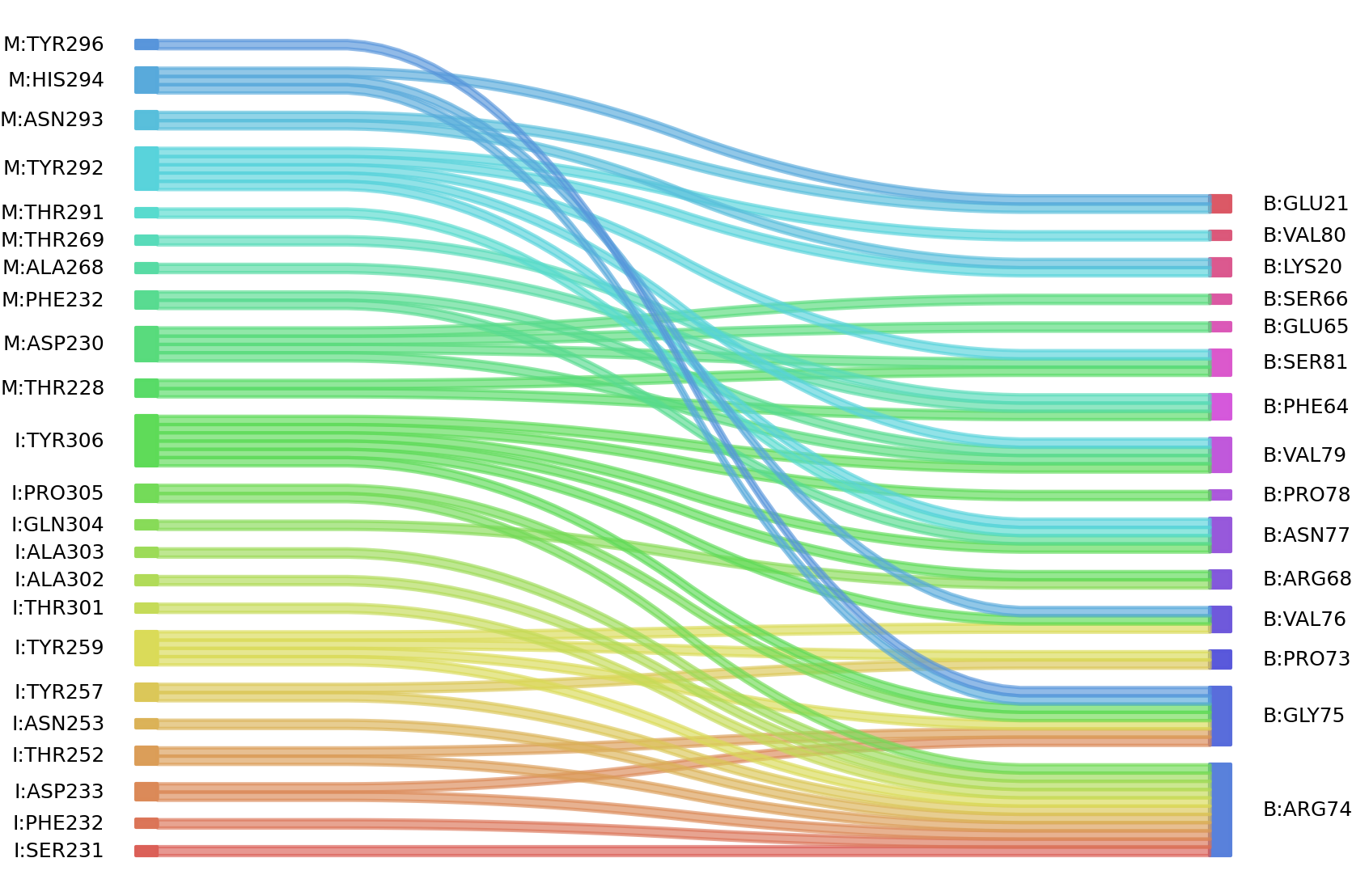
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition M: SER226 THR228 ASP230 PHE232 ALA268 THR269 THR291 TYR292 ASN293 HIS294 TYR296 I: SER231 ASP233 SER250 THR252 ASN253 TYR257 TYR259 GLN299 ALA302 ALA303 GLN304 PRO305 TYR306 B: LYS20 GLU21 PHE64 SER66 ARG68 PRO73 ARG74 GLY75 VAL76 ASN77 PRO78 VAL79 VAL80 SER81 THR97 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)