Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 136 |
| PDBID: | 1P2C |
| Chains: | ED_F |
| Organism: | Mus musculus, Gallus gallus |
| Method: | XRD |
| Resolution (Å): | 2.00 |
| Reference: | 10.1073/pnas.0400060101 |
| Antibody | |
| Antibody: | F10.6.6 Fab |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | Hen Egg White Lysozyme |
| Antigen mutation: | No |
| Durg Target: | |
Sequence information
Antibody
Heavy Chain: E
Mutation: NULL
| >1P2C_E|Chain B, E|heavy chain VH+CH1 anti-lysozyme antibody F10.6.6|Mus musculus (10090) EVQLQESGAELMKPGASVKISCKATGYTFTTYWIEWIKQRPGHSLEWIGEILPGSDSTYYNEKVKGKVTFTADASSNTAYMQLSSLTSEDSAVYYCARGDGFYVYWGQGTTLTVSSASTTPPSVYPLAPGSAAQTNSMVTLGCLVKGYFPEPVTVTWNSGSLSSGVHTFPAVLQSDLYTLSSSVTVPSSPWPSETVTCNVAHPASSTKVDKKIVPRDC |
Light Chain: D
Mutation: NULL
| >1P2C_D|Chain A, D|light chain anti-lysozyme antibody F10.6.6|Mus musculus (10090) DIELTQSPATLSVTPGDSVSLSCRASQSISNNLHWYQQKSHESPRLLIKYTSQSMSGIPSRFSGSGSGTDFTLSINSVETEDFGVYFCQQSGSWPRTFGGGTKLDIKRADAAPTVSIFPPSSEQLTSGGASVVCFLNNFYPKDINVKWKIDGSERQNGVLNSWTDQDSKDSTYSMSSTLTLTKDEYERHNSYTCEATHKTSTSPIVKSFNRN |
Antigen
Chain: F
Mutation: NULL
| >1P2C_F|Chain C, F|Lysozyme C|Gallus gallus (9031) KVFGRCELAAAMKRHGLDNYRGYSLGNWVCAAKFESNFNTQATNRNTDGSTDYGILQINSRWWCNDGRTPGSRNLCNIPCSALLSSDITASVNCAKKIVSDGNGMNAWVAWRNRCKGTDVQAWIRGCRL |
Interaction
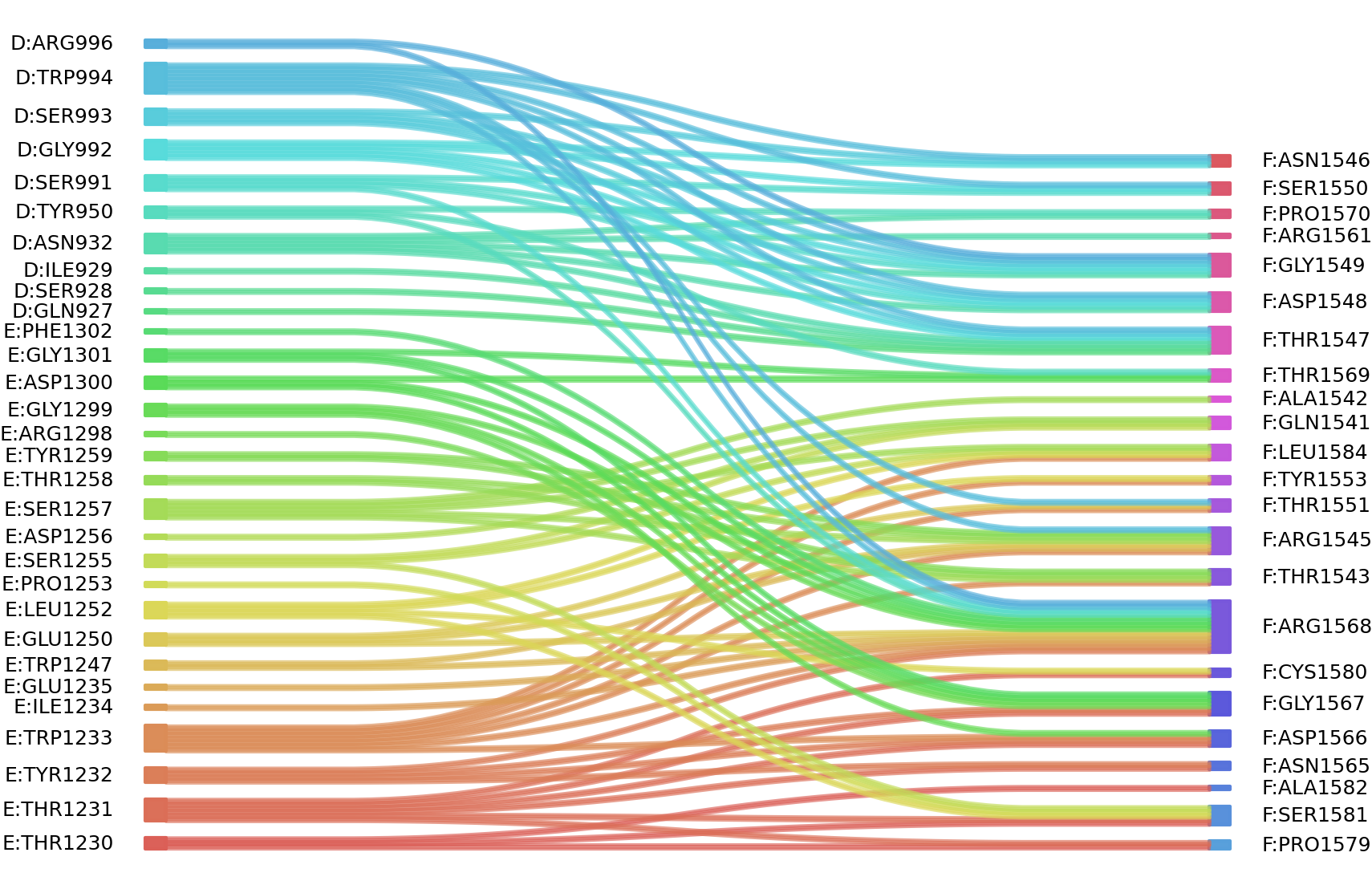
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition D: GLN927 SER928 ILE929 ASN932 TYR950 SER991 GLY992 SER993 TRP994 ARG996 E: THR1230 THR1231 TYR1232 TRP1233 GLU1235 GLU1250 LEU1252 SER1255 ASP1256 SER1257 THR1258 TYR1259 GLY1299 ASP1300 GLY1301 F: GLN1541 THR1543 ARG1545 ASN1546 THR1547 ASP1548 GLY1549 SER1550 THR1551 TYR1553 ASN1565 ASP1566 GLY1567 ARG1568 PRO1570 PRO1579 SER1581 LEU1584 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)