Details
Structure visualisation
Entry information
| Complex | |
| AACDB_ID: | 1312 |
| PDBID: | 5F72 |
| Chains: | S_K |
| Organism: | Homo sapiens |
| Method: | XRD |
| Resolution (Å): | 1.85 |
| Reference: | Not available or To be published |
| Antibody | |
| Antibody: | 5F72 scFv |
| Antibody mutation: | No |
| INN (Clinical Trial): | |
| Antigen | |
| Antigen: | Kelch-like ECH-associated protein 1 |
| Antigen mutation: | No |
| Durg Target: | Q14145 |
Sequence information
Antibody
Chain: S
Mutation: NULL
| >5F72_S|Chain C[auth S], D[auth T]|Single chain Fv from a Fab|Homo sapiens (9606) EVQLVESGGGLVQPGGSLRLSCAASGFAISASSIHWVRQAPGKCLEWVASIDPETGETLYAKSVAGRFTISADTSKNTAYLQMNSLRAEDTAVYYCARAYAGDGVYYADVWGQGTLVTVSSGGGGSGGGGSGGGGSGGGGSDIQMTQSPSSLSASVGDRVTITCRASQSVSSAVAWYQQKPGKAPKLLIYSASSLYSGVPSRFSGSRSGTDFTLTISSLQPEDFATYYCQQSYSFPSTFGCGTKVEIKRTENLYFQG |
Antigen
Chain: K
Mutation: NULL
| >5F72_K|Chain A[auth C], B[auth K]|Kelch-like ECH-associated protein 1|Homo sapiens (9606) GSMGHAPKVGRLIYTAGGYFRQSLSYLEAYNPSDGTWLDLADLQVPRSGLAGCVVGGLLYAVGGRNNSPDGNTDSSALDCYNPMTNQWSPCAPMSVPRNRIGVGVIDGHIYAVGGSHGCIHHNSVERYEPERDEWHLVAPMLTRRIGVGVAVLNRLLYAVGGFDGTNRLNSAECYYPERNEWRMITAMNTIRSGAGVCVLHNCIYAAGGYDGQDQLNSVERYDVETETWTFVAPMKHRRSALGITVHQGRIYVLGGYDGHTFLDSVECYDPDTDTWSEVTRMTSGRSGVGVAVTME |
Interaction
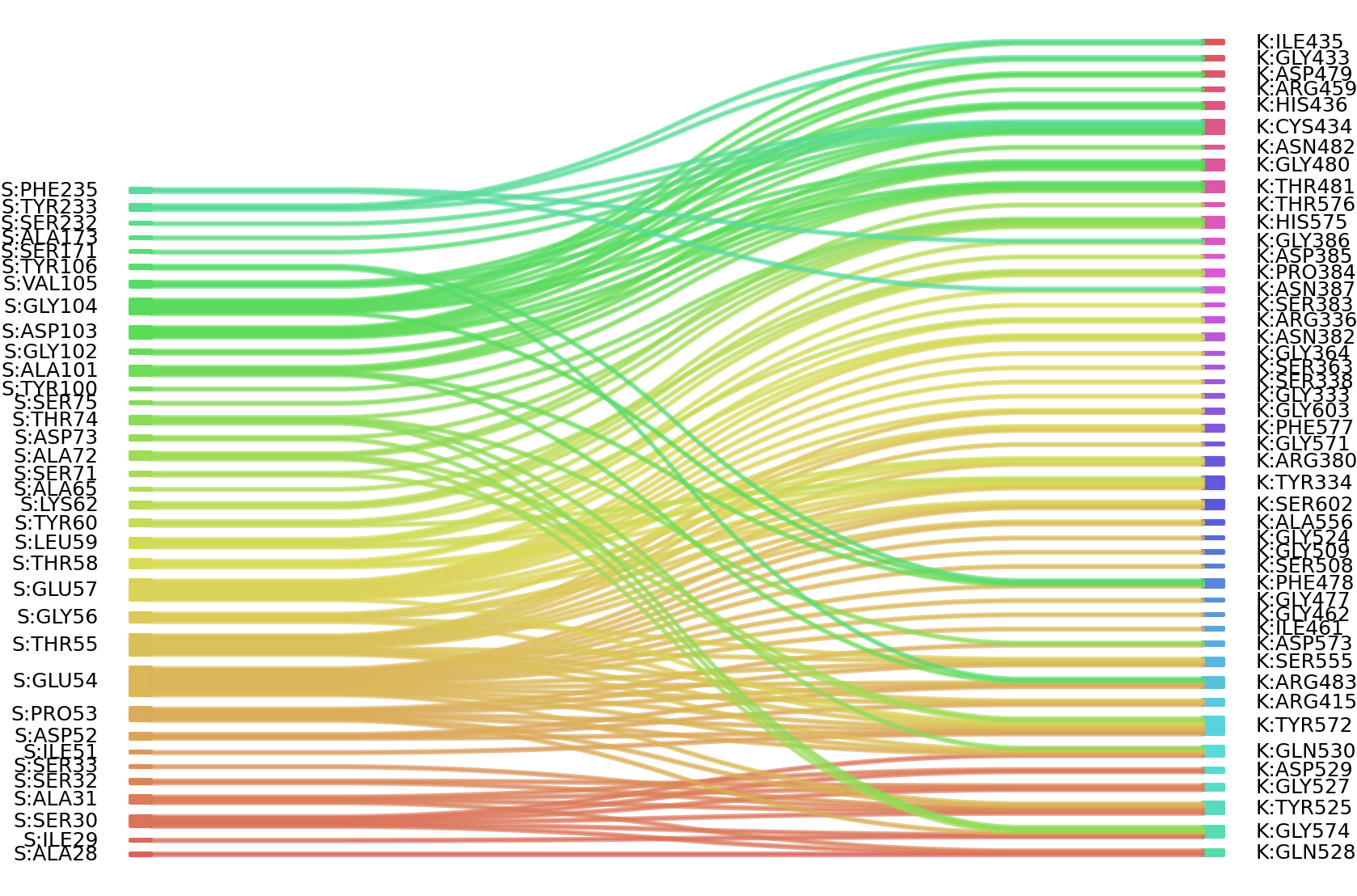
1、Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1) for each residue in antibody and antigen, respectively. The residues with SASA loss in binding of more than 1Å2 were classified as interacting residues.
Interacting residues (ΔSASA based)
| Chain residues position delta_SASA
: residuesposition S: ALA28 SER30 ALA31 SER32 ILE51 ASP52 PRO53 GLU54 THR55 GLY56 GLU57 THR58 LEU59 TYR60 LYS62 ALA65 GLY66 SER71 ALA72 ASP73 THR74 SER75 TYR100 ALA101 GLY102 ASP103 GLY104 VAL105 TYR106 SER171 ALA173 SER232 TYR233 PHE235 K: TYR334 ARG336 SER363 ARG380 ASN382 PRO384 ASP385 GLY386 ASN387 ARG415 GLY433 CYS434 ILE435 HIS436 ARG459 PHE478 ASP479 GLY480 THR481 ARG483 SER508 TYR525 GLY527 GLN528 ASP529 GLN530 SER555 ALA556 TYR572 ASP573 GLY574 HIS575 PHE577 SER602 GLY603 |
2、We defined interacting paratope-epitope residues by a distance cutoff of < 6 Å . Two amino acids are considered as interacting residues if they have at least one atom within a distance of 6 Å from any atom.
Interacting residues (Atom distance based)