Background
BDB is short for Biopanning Data Bank. It is a special database collecting peptides that have been selected from random peptide libraries based on their ability to bind various substances ranging from small compounds to whole organisms through surface display technology especially phage display technology. It aims to be an information portal to experimental biopanning data. Besides the peptide sequences, other information such as the corresponding target, template, library, and structures are also stored. All entries are manually extracted from published peer review articles and other public data sources such as Uniprot, GenBank and PDBSum.
Phages are short for bacteriophages. They are viruses that infect bacteria. Many phages such as M13 and fd are good expression vectors. In 1985, George P. Smith first described phage display, when he displayed foreign peptides on the virion surface by inserting the foreign DNA fragments into the filamentous phage gene III [1]. He also demonstrated that foreign peptides in fusion proteins on the virion surface were in immunologically accessible form and specific fusion phage could be enriched and isolated from a phage library of random inserts in a fusion-phage vector by one or more rounds of affinity selection. Subsequently, other surface display systems such as bacterial display (1986) [2], ribosome display (1994) [3], mRNA display (1997) [4] and yeast display (1997) [5] developed one after another.
Usually, the substance used to screen random peptide library is termed target. The screening of the library with an intended target (bait) is called biopanning or panning in short [6]. As an artificial selection and efficient evolution technique in vitro or in vivo, biopanning has been widely used in mapping epitopes [7], defining protein interaction networks [8], and developing new diagnostics [9, 10], therapeutics [11, 12] and vaccines [13]. Therefore, biopanning data is valuable and deserves a special database, especially when they are not collected in main stream resources at NCBI and EBI [14].
1.2 Mimotope and TUP (Target-unrelated Peptide)
Peptide mimicking the binding site on the template and capable of binding to the target is defined as mimotope, which was first introduced by Mario Geysen et al [15]. In the biopanning result however, there are not only mimotopes (desired signal), but also all kinds of target-unrelated peptides (TUPs) (unwanted noise) [16, 17]. TUPs can be divided into two categories [16]. One category of TUP is called selection-related TUP (SrTUP). Although unable to bind to the target site, they can react with contaminants or other components of the screening system and then sneak into the biopanning results. Another category of TUP is called propagation-related TUP (PrTUP). They creep into the output of biopanning because they have a higher infection rate or faster secretion rate [18, 19].
In 2010, we developed the MimoDB database (http://immunet.cn/mimodb/) and aimed to provide a repository for mimotope data derived from phage display technology [20]. Since MimoDB 2.0, panning data from mRNA display, ribosome display, bacterial display and yeast display have also been curated and we aim to build MimoDB into an information portal to biopanning results of random peptide libraries [21]. From the release of September, 2014, the database was renamed as BDB. The original name MimoDB is, in fact, an abbreviation of two separate words: mimotope database. At that time we initiated MimoDB, we thought all peptides obtained by biopanning should be mimotopes. Therefore, we coined this name. However, the fact is that biopanning data is usually a mixture of mimotopes and TUPs. To avoid any possible misleading or confusion caused by the original name, we rebrand the database to BDB which stands for Biopanning Data Bank.
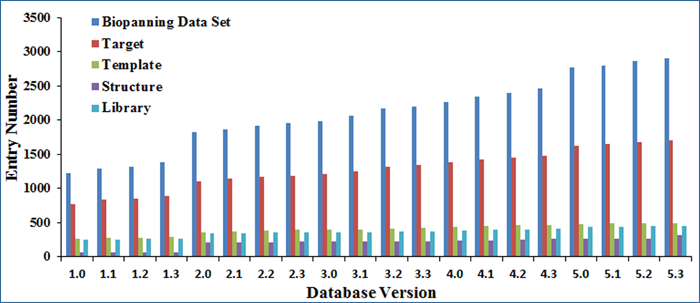
The BDB database is usually revised and updated quarterly. It has been updated 19 times since its first release and 15 times since version 2.0 (August 21, 2011 release). The current version (version 5.3) is July 22, 2015 release, which contains 2,904 sets of biopanning data collected from 1,322 papers, including 25,786 peptide sequences, 1,704 targets, 492 known templates, 447 peptide libraries, and 310 crystal structures of target-template or target-peptide complex. As shown below, the biopanning data sets and relevant data entries have increased substantially compared with those of version 2.0 .
Explanations for basic terms in BDB.
Phage | A category of viruses that infect bacteria. Some phages are good expression vector to accommodate foreign DNA and express the foreign DNA as an insert peptide in a fusion protein. In case of the phage coat protein, the foreign peptide is displayed on the outer surface of the phage particles. |
Phage display | An in vitro methodology and system for presenting, selecting and evolving proteins and peptides displayed on the surface of phage particles. |
Yeast display | An in vitro methodology and system for presenting, selecting and evolving proteins and peptides displayed on the surface of yeast. |
Bacterial display | A protein engineering technique for presenting, selecting and evolving proteins and polypeptides on the surface of bacteria. |
Ribosome display | An in vitro methodology and system for generating, selecting, and evolving proteins and peptides. Its key idea is to translate a library of mRNA molecules with a stoichiometric quantity of ribosomes. |
Phage library | A mixture of phage clones with each carrying a different foreign DNA insert and therefore displaying a different peptide on its surface. The library complexity is the estimated number of unique phage clones in a library. The library titer is a measure of the concentration of infectious phage in a library, usually the number of plaque forming units per mL. This number can then be used to dilute the phage allowing maximum plaques without plaque superimposition. |
Target | The substance used to screen the random peptide library. The target nearly can be anything, varying from small compounds to nucleic acids, proteins, cells, tissues, organs and even entire organisms. Nonetheless, proteins including antibodies and receptors from human and mouse are the most used targets. |
Template | The genuine partner mimicked by mimotopes and binding the target. However, sometimes the template may not exist or can not be determined. For example, it is hard to assign a template for the peptides selected with plastic. On one hand, it is quite often that the partner of a target is unknown. On the other hand, a target may bind many partners and a lot of papers do not have enough data to determine which one is the template for a mimotope. |
Mimotope | Peptide mimicking the binding site on the template and binding to the target. Whether a peptide is mimotope or target-unrelated peptide is dependent on the target used in experiment. For example, an Fc-binding peptide is a target-unrelated peptide when Fab fragments are used to select phage library; however, the same peptide is a mimotope when using Fc fragment as target. |
Target-unrelated peptide | Peptide reacting with contaminant or other components of the screening system rather than binding to the target site, or peptide with higher infection rate or faster secretion rate. |
Biopanning | Screening of the library with an intended target (bait). Each round of panning consists of two basic processes: selection and amplification. |
| Biopanning Data | Selected peptides through iterative rounds of biopanning. |
Reference
1. Smith, G.P. (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science, 228, 1315-1317.
2. Freudl, R., MacIntyre, S., Degen, M. and Henning, U. (1986) Cell surface exposure of the outer membrane protein OmpA of Escherichia coli K-12. Journal of molecular biology, 188, 491-494.
3. Mattheakis, L.C., Bhatt, R.R. and Dower, W.J. (1994) An in vitro polysome display system for identifying ligands from very large peptide libraries. Proceedings of the National Academy of Sciences of the United States of America, 91, 9022-9026.
4. Roberts, R.W. and Szostak, J.W. (1997) RNA-peptide fusions for the in vitro selection of peptides and proteins. Proceedings of the National Academy of Sciences of the United States of America, 94, 12297-12302.
5. Boder, E.T. and Wittrup, K.D. (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nature biotechnology, 15, 553-557.
6. Ehrlich, G.K., Berthold, W. and Bailon, P. (2000) Phage display technology. Affinity selection by biopanning. Methods in molecular biology, 147, 195-208.
7. He, B., Mao, C., Ru, B., Han, H., Zhou, P. and Huang, J. (2013) Epitope mapping of metuximab on CD147 using phage display and molecular docking. Computational and mathematical methods in medicine, 2013, 983829.
8. Tong, A.H., Drees, B., Nardelli, G., Bader, G.D., Brannetti, B., Castagnoli, L., Evangelista, M., Ferracuti, S., Nelson, B., Paoluzi, S. et al. (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science, 295, 321-324.
9. Larimer, B.M. and Deutscher, S.L. (2014) Development of a peptide by phage display for SPECT imaging of resistance-susceptible breast cancer. American journal of nuclear medicine and molecular imaging, 4, 435-447.
10. Stewart, L.D., Foddai, A., Elliott, C.T. and Grant, I.R. (2013) Development of a novel phage-mediated immunoassay for the rapid detection of viable Mycobacterium avium subsp. paratuberculosis. Journal of applied microbiology, 115, 808-817.
11. Vodnik, M., Molek, P., Strukelj, B. and Lunder, M. (2013) Peptides binding to the hunger hormone ghrelin. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme, 45, 372-377.
12. Yang, M., Liu, C., Niu, M., Hu, Y., Guo, M., Zhang, J., Luo, Y., Yuan, W., Yang, M., Yun, M. et al. (2014) Phage-display library biopanning and bioinformatic analysis yielded a high-affinity peptide to inflamed vascular endothelium both in vitro and in vivo. Journal of controlled release : official journal of the Controlled Release Society, 174, 72-80.
13. Bachler, B.C., Humbert, M., Palikuqi, B., Siddappa, N.B., Lakhashe, S.K., Rasmussen, R.A. and Ruprecht, R.M. (2013) Novel biopanning strategy to identify epitopes associated with vaccine protection. Journal of virology, 87, 4403-4416.
14. Huang, J., Ru, B. and Dai, P. (2011) Bioinformatics resources and tools for phage display. Molecules, 16, 694-709.
15. Geysen, H.M., Rodda, S.J. and Mason, T.J. (1986) A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Molecular immunology, 23, 709-715.
16. Vodnik, M., Zager, U., Strukelj, B. and Lunder, M. (2011) Phage display: selecting straws instead of a needle from a haystack. Molecules, 16, 790-817.
17. Matochko, W.L., Cory Li, S., Tang, S.K. and Derda, R. (2014) Prospective identification of parasitic sequences in phage display screens. Nucleic acids research, 42, 1784-1798.
18. Brammer, L.A., Bolduc, B., Kass, J.L., Felice, K.M., Noren, C.J. and Hall, M.F. (2008) A target-unrelated peptide in an M13 phage display library traced to an advantageous mutation in the gene II ribosome-binding site. Analytical biochemistry, 373, 88-98.
19. Thomas, W.D., Golomb, M. and Smith, G.P. (2010) Corruption of phage display libraries by target-unrelated clones: diagnosis and countermeasures. Analytical biochemistry, 407, 237-240.
20. Ru, B., Huang, J., Dai, P., Li, S., Xia, Z., Ding, H., Lin, H., Guo, F. and Wang, X. (2010) MimoDB: a new repository for mimotope data derived from phage display technology. Molecules, 15, 8279-8288.
21. Huang, J., Ru, B., Zhu, P., Nie, F., Yang, J., Wang, X., Dai, P., Lin, H., Guo, F.B. and Rao, N. (2012) MimoDB 2.0: a mimotope database and beyond. Nucleic acids research, 40, D271-277.