Search
At the front-end, five tables, (BiopanningDataSet, Target, Template, Library and Structure), can be searched. One could choose the simple search or advanced search, enter their keywords and press search button to start search. A paged summary table would appear at first. Search results can be downloaded as xml files by clicking on the "Download this result" or "Download selected" option. If you chose "Download this result", all results would be downloaded. Results that were checked would be downloaded if you clicked on the “Download selected” option.
Users can select the simple search to search each table separately. The simple search bar is showed below.

If a user chose the "Everything" tab, searching would be performed in five tables, respectively. The following fields can be used to search BDB.
Sequence (All sequences are recorded in one-letter amino acid codes in BDB.)
Experimental Method (Common panning: eluting with acid buffer. Subtractive panning: with additional negative selection steps to remove non-specific binder. Competitive panning: eluting with template or target. In vivo panning: injecting peptide library into the model organism.)
Reference (PMID if recorded in PubMed, otherwise the DOI name or URL.)
Target Name (Name of the target. The recommended name of UniProt is used if available.)
Target Source (The organism name to which the target belongs. For some targets, such as small compounds and macromolecular materials (e.g. cellulose and polystyrene), this field is null. )
Target Type (All targets are grouped into following types: organ and tissue, cell, protein (monoclonal antibody, polyclonal antibody, receptor, others), nucleic acid, inorganic materials and miscellaneous.)
Template Name (Name of the template. To standardize template name, the recommended name of UniProt is used if available)
Template Source (The organism name to which the template belongs. For some targets, such as small compounds and macromolecular materials (e.g. cellulose and polystyrene), this field is null.)
Template Type (All templates are grouped into following types: organ and tissue, cell, protein (monoclonal antibody, polyclonal antibody, receptor, others), nucleic acid, inorganic materials and miscellaneous.)
Library Name (Name of the library such as Ph.D.-12 phage display library, Ph.D.-7 phage display library and Ph.D.-C7C phage display library (CX7C).)
PDB ID (PDB code of the complex structure.)
For example, if one wanted to retrieve biotin in all five tables, you would first select the "Everything" tab, input "biotin" into the blank text area and then click on the "Search" tab. Finally, you could see the result page and a subset of the 5 records returned from a query using biotin was shown. The search results could be downloaded as xml files. If you wished to see the detailed information, you could choose the corresponding IDs or Names to view the detailed information. The Corresponding Entries field summarizes how many sets of biopanning data which include this target or template and can be hyperlinked to those biopanning data sets.

Clicking on the "45 Entries" in above table, you could see 45 sets of biopanning data, in which biotin serves as template. These related biopanning data sets can be downloaded by clicking on the “Download all entries” or “Download selected” option.
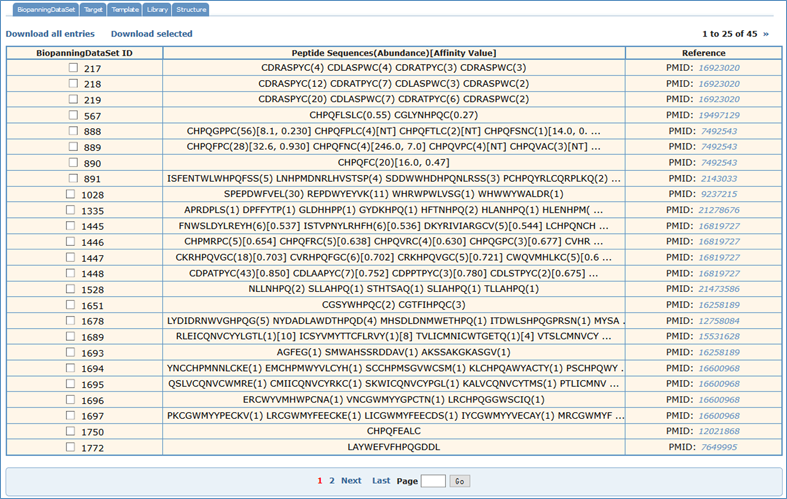
For searching the BiopanningDataSet table, users should choose the "BiopannningDataSet" tab. Four fields (BiopannningDataSet ID, Sequences, Experimental Method and Reference) can be searchable. For example, if one wanted to retrieve all biopanning data sets which contain CSWRPPFRAVC, you would first select the "BiopanningDataSet" tab, input "CSWRPPFRAVC" into the blank text area and then click on the "Search" tab. Finally, you could see the search results in a paged summary table, in which are the fields of BiopanningDataSet ID, Peptide Sequences(Abundance)[Affinity Value] and Reference. This field lists part or all of peptides in a set of biopanning data with peptide abundance and affinity value. The Reference field lists PMID if recorded in PubMed, otherwise the DOI name or URL and can be hyperlinked to corresponding manuscript in other public sources.

To search the Target table, users should select the "Target" tab. Four fields (Target ID, Target Name, Target Source and Target Type) can be searched. For example, if one wanted to retrieve all targets which contain interleukin, you would first select the "Target" tab, input "interleukin" into the blank text area and then click on the "Search" tab. Finally, you could see the result page as shown below. If you wished to see the detailed information, you could choose the corresponding IDs or Names to view the detailed information. The Corresponding Entries field summarizes how many sets of biopanning data which include this target and can be hyperlinked to those biopanning data sets.

For searching the Template table, one could choose the "Template" tab. Four fields (Template ID, Template Name, Template Source and Template Type) can be searched. For example, if one wanted to retrieve all templates which contain surface protein gp120, you would first select the "Template" tab, input "surface protein gp120" and then click on the "Search" tab. Finally, you could see the result page as shown below. If you wished to see the detailed information, you could choose the corresponding IDs or Names to view the detailed information. The Corresponding Entries field summarizes how many sets of biopanning data which include this template and can be hyperlinked to those biopanning data sets.

To search for a peptide library, one had better choose the "Library" tab. The fields, Library ID and Library Name, can be used to search the Library table. For example, if one wanted to retrieve Ph.D.-12 phage display library, you would first select the "Library" tab, input "Ph.D.-12 phage display library" and then click on the "Search" tab. Finally, you could see the result page as shown below. If you wished to see the detailed information, you could choose the corresponding IDs or Names to view the detailed information. The Corresponding Entries field summarizes how many sets of biopanning data which include this library and can be hyperlinked to those biopanning data sets.

In seek of a structure, the "Structure" tab is a preferable choice. PDB code of the complex structure or Complex ID can be used to search. For example, if one wanted to retrieve 1G1S, you would first select the "Structure" tab, input "1G1S" and then click on the "Search" tab. Finally, you could see the result page as shown below. If you wished to see the detailed information, you could choose the corresponding IDs or Names to view the detailed information. The Corresponding Entries field summarizes how many sets of biopanning data which include this complex structure and can be hyperlinked to those biopanning data sets.

BDB can be searched by most fields in the 5 main tables and by any combination of up to three of the fields. There are three pull-down list boxes and two pairs of radio buttons on the advanced search interface. A query is formed by selecting a table from the pull-down list box and filling some fields in the table. Any words, strings and numbers such as peptide sequence, protein name, species name, PDB code, PMID of reference and etc. can be entered into the blank text forms. For the option "PDBID, Null, Not Null" of the target-template complex option group in the list box, two special words, i.e. null and not null are supported, which mean with or without solved structure of target-template complex.
For example, if one wanted to retrieve all biopanning data sets selected with monoclonal antibodies against protein antigens and at the same time the structures of corresponding antibody-antigen complexes have been solved, you would first select the search field "Target type" from the pull-down list box, input "monoclonal antibody" into the corresponding blank text form and then select the "and" button. Then you would select the "Template Type" search field and input "protein". Finally, you would select "PDBID, Null, and Not Null" field from the target-template complex option group and input" not null".

Search results can be downloaded in xml files by clicking "Download this result" or "Download Selected".
